关于使用Scipy处理lognorm分布(docs),已经有相当多的帖子,但我仍然不太明白。
对数正态分布通常由两个参数\mu和\sigma描述,这对应于Scipy参数loc=0和\sigma=shape,\mu=np.log(scale)。
在scipy, 对数正态分布-参数,我们可以了解到如何使用随机分布的指数生成一个lognorm(\mu,\sigma)样本。现在让我们试试其他方法:
A)
直接创建lognorm存在什么问题:
import scipy as sp
import matplotlib.pyplot as plt
# lognorm(mu=10,sigma=3)
# so shape=3, loc=0, scale=np.exp(10) ?
x=np.linspace(0.01,20,200)
sample_dist = sp.stats.lognorm.pdf(x, 3, loc=0, scale=np.exp(10))
shape, loc, scale = sp.stats.lognorm.fit(sample_dist, floc=0)
print shape, loc, scale
print np.log(scale), shape # mu and sigma
# last line: -7.63285693379 0.140259699945 # not 10 and 3
B)
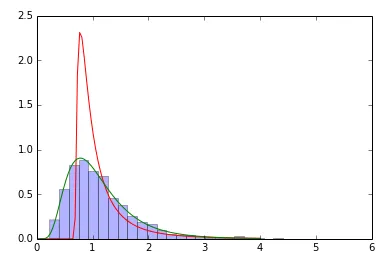
我使用拟合的返回值来创建拟合分布。但是显然我又做错了什么:
samp=sp.stats.lognorm(0.5,loc=0,scale=1).rvs(size=2000) # sample
param=sp.stats.lognorm.fit(samp) # fit the sample data
print param # does not coincide with shape, loc, scale above!
x=np.linspace(0,4,100)
pdf_fitted = sp.stats.lognorm.pdf(x, param[0], loc=param[1], scale=param[2]) # fitted distribution
pdf = sp.stats.lognorm.pdf(x, 0.5, loc=0, scale=1) # original distribution
plt.plot(x,pdf_fitted,'r-',x,pdf,'g-')
plt.hist(samp,bins=30,normed=True,alpha=.3)