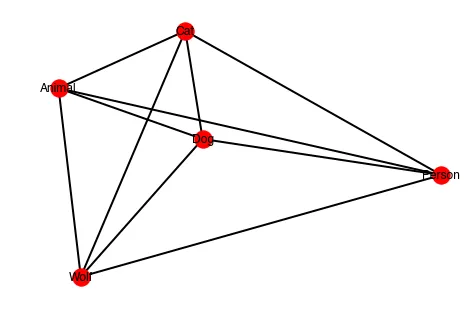
我正在尝试使用networkx可视化单词之间的相关性(相似度得分高达1)
例如,狗、猫、动物、人、狼之间的相似性得分
我已经尝试使用以下代码绘制每个单词/节点之间的相似性距离
import networkx as nx
import matplotlib.pyplot as plt
G=nx.Graph()
corr_data =([['Dog', 'Dog', 1.0],
['Cat', 'Dog', 0.8016854524612427],
['Wolf', 'Dog', 0.5206573009490967],
['Person', 'Dog', 0.3756750822067261],
['Animal', 'Dog', 0.6618534326553345],
['Cat', 'Cat', 1.0],
['Wolf', 'Cat', 0.5081626176834106],
['Person', 'Cat', 0.32475101947784424],
['Animal', 'Cat', 0.6260400414466858],
['Wolf', 'Wolf', 1.0],
['Person', 'Wolf', 0.23091702163219452],
['Animal', 'Wolf', 0.5261368751525879],
['Person', 'Person', 1.0],
['Animal', 'Person', 0.34220656752586365],
['Animal', 'Animal', 1.0]])
existing_edges = {}
def build_graph(w, lev):
if (lev > 5) :
return
for z in corr_data:
ind=-1
if z[0] == w:
ind=0
ind1=1
if z[1] == w:
ind ==1
ind1 =0
if ind == 0 or ind == 1:
if str(w) + "_" + str(corr_data[ind1]) not in existing_edges :
G.add_node(str(corr_data[ind]))
existing_edges[str(w) + "_" + str(corr_data[ind1])] = 1;
G.add_edge(w,str(corr_data[ind1]))
build_graph(corr_data[ind1], lev+1)
existing_nodes = {}
def build_graph_for_all():
count=0
for d in corr_data:
if (count > 40) :
return
if d[0] not in existing_edges :
G.add_node(str(d[0]))
if d[1] not in existing_edges :
G.add_node(str(d[1]))
G.add_edge(str(d[0]), str(d[1]))
count=count + 1
build_graph_for_all()
print (G.nodes(data=True))
plt.show()
nx.draw(G, width=2, with_labels=True)
plt.savefig("path1.png")
w="design"
G.add_node(w)
build_graph(w, 0)
print (G.nodes(data=True))
plt.show()
nx.draw(G, width=2, with_labels=True)
plt.savefig("path.png")
我的节点之间的距离似乎出现了问题,例如猫和人的距离比猫和狗的距离更近。我是否忽略了明显的问题?