我正在使用scipy的gaussian_kde来获取一些双峰数据的概率密度。但是,由于我的数据是角度(以度数表示的方向),当值接近极限时,我遇到了问题。下面的代码给出了两个示例kde,在定义域为0-360时会低估,因为它无法处理数据的循环性质。pdf需要在单位圆上定义,但我找不到任何适用于这种类型数据的scipy.stats函数(von mises分布存在,但仅适用于单峰数据)。有人遇到过这个问题吗?是否有任何(最好基于python)可用于估计单位圆上的双峰pdf?
import numpy as np
import scipy as sp
from pylab import plot,figure,subplot,show,hist
from scipy import stats
baz = np.array([-92.29061004, -85.42607874, -85.42607874, -70.01689348,
-63.43494882, -63.43494882, -70.01689348, -70.01689348,
-59.93141718, -63.43494882, -59.93141718, -63.43494882,
-63.43494882, -63.43494882, -57.52880771, -53.61564818,
-57.52880771, -63.43494882, -63.43494882, -92.29061004,
-16.92751306, -99.09027692, -99.09027692, -16.92751306,
-99.09027692, -16.92751306, -9.86580694, -8.74616226,
-9.86580694, -8.74616226, -8.74616226, -2.20259816,
-2.20259816, -2.20259816, -9.86580694, -2.20259816,
-2.48955292, -2.48955292, -2.48955292, -2.48955292,
4.96974073, 4.96974073, 4.96974073, 4.96974073,
-2.48955292, -2.48955292, -2.48955292, -2.48955292,
-2.48955292, -9.86580694, -9.86580694, -9.86580694,
-16.92751306, -19.29004622, -19.29004622, -26.56505118,
-19.29004622, -19.29004622, -19.29004622, -19.29004622])
xx = np.linspace(-180, 180, 181)
scipy_kde = stats.gaussian_kde(baz)
print scipy_kde.integrate_box_1d(-180,180)
figure()
plot(xx, scipy_kde(xx), c='green')
baz[baz<0] += 360
xx = np.linspace(0, 360, 181)
scipy_kde = stats.gaussian_kde(baz)
print scipy_kde.integrate_box_1d(-180,180)
plot(xx, scipy_kde(xx), c='red')
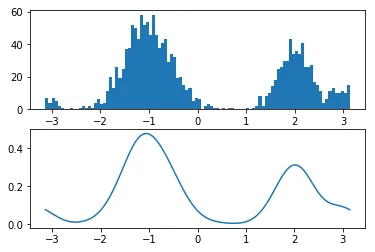
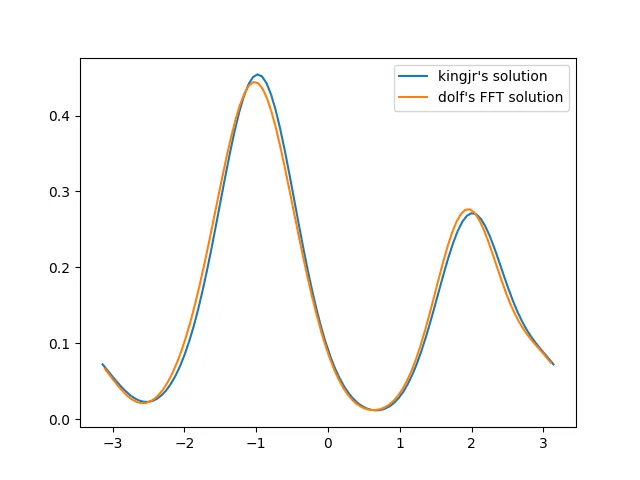
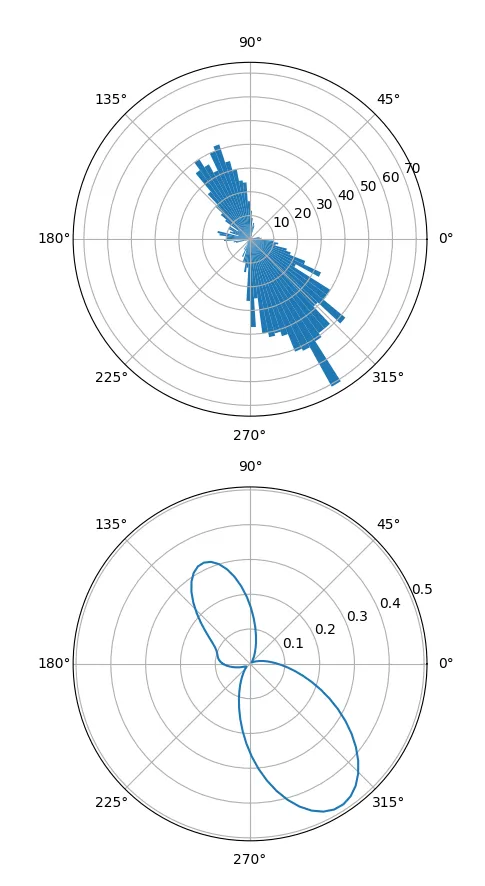
def vonmises_kde...中,您可以省去一个变量x或bins。 - Dzamo Norton