早上好!根据我所找到的信息,您可以按照以下方式使用ranger()和fastshap():
library(fastshap)
library(ranger)
library(tidyverse)
data(iris)
df <- iris
df$Target <- if_else(df$Species == "setosa",1,0)
df$Species <- NULL
x <- df %>% select(-Target)
model <- ranger(
x = df %>% select(-Target),
y = df %>% pull(Target))
pfun <- function(object, newdata) {
predict(object, data = newdata)$predictions
}
system.time({
set.seed(5038)
shap <- fastshap::explain(model, X = x, pred_wrapper = pfun, nsim = 10)
})
library(ggplot2)
theme_set(theme_bw())
shap_imp <- data.frame(
Variable = names(shap),
Importance = apply(shap, MARGIN = 2, FUN = function(x) sum(abs(x)))
)
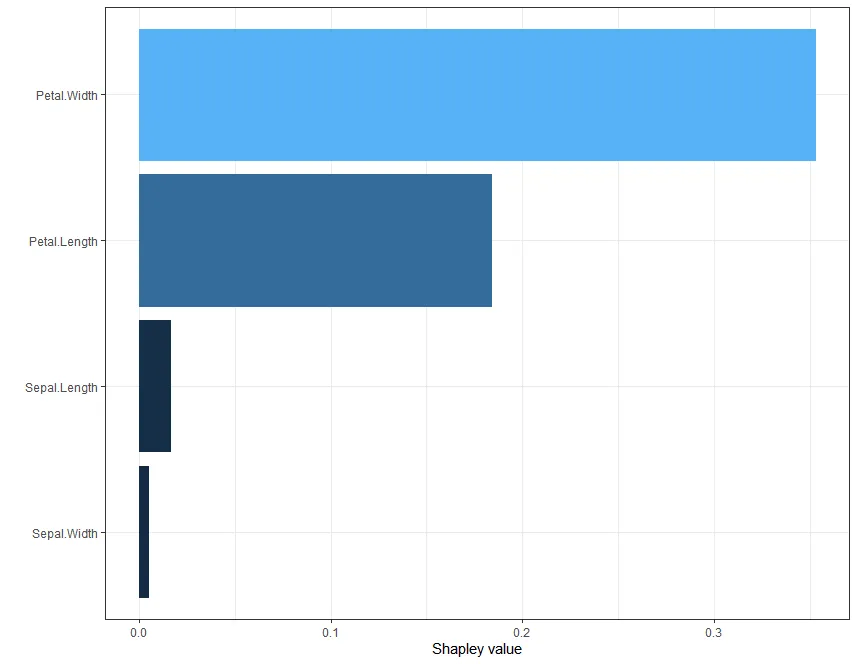
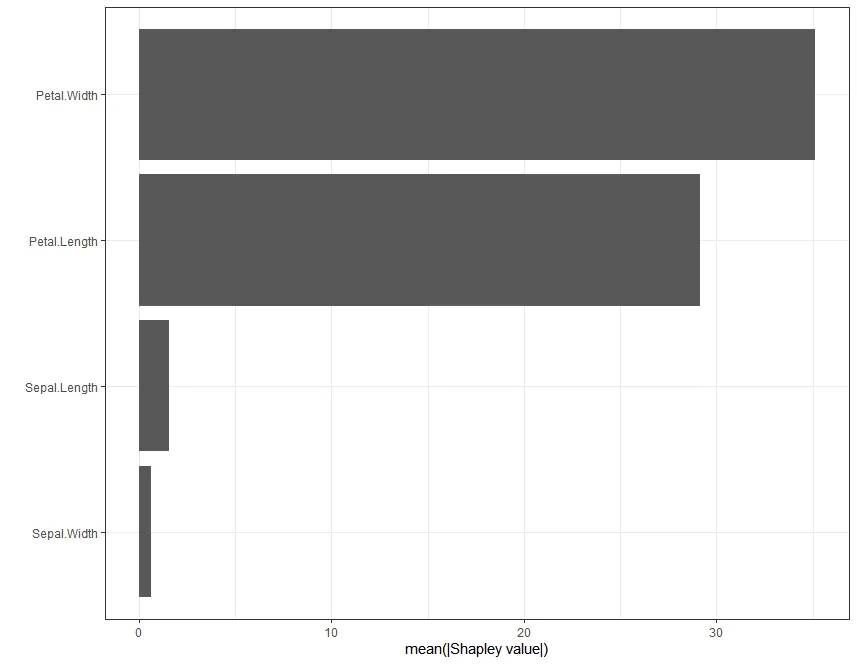
例如,对于变量重要性,您可以进行以下操作:
# Plot Shap-based variable importance
ggplot(shap_imp, aes(reorder(Variable, Importance), Importance)) +
geom_col() +
coord_flip() +
xlab("") +
ylab("mean(|Shapley value|)")
如果你想获得个别预测,可以采用以下方式:
expl <- fastshap::explain(model, X = x ,pred_wrapper = pfun, nsim = 10, newdata = x[1L, ])
autoplot(expl, type = "contribution")
所有这些信息都可以在这里找到,还有更多的内容:
https://bgreenwell.github.io/fastshap/articles/fastshap.html
点击链接查看并解决您的疑惑! :)