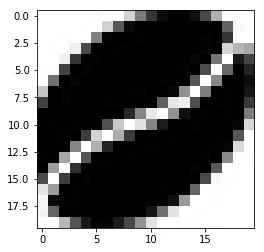
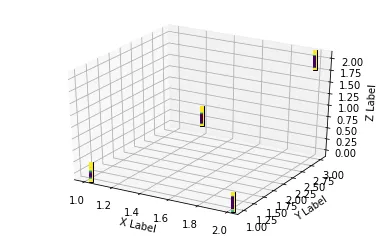
这里有一个不太正式的解决方案,它将图像转换为数据框,其中每个像素变成一个体素(?),然后我们将其发送到plotly中。它基本上可以工作,但需要一些改进来:
1) 更多地调整图像(使用腐蚀步骤?)以排除更多低alpha像素
2) 在plotly中使用请求的颜色范围
步骤1:导入图像并调整大小,并过滤掉透明或部分透明的像素
library(tidyverse)
library(magick)
sprite_frame <- image_read("coffee-bean-for-a-coffee-break.png") %>%
magick::image_resize("20x20") %>%
image_raster(tidy = T) %>%
mutate(alpha = str_sub(col, start = 7) %>% strtoi(base = 16)) %>%
filter(col != "transparent",
alpha > 240)
编辑:如果这个代码块对任何人有用,我会添加结果:
sprite_frame <-
structure(list(x = c(13L, 14L, 10L, 11L, 12L, 13L, 14L, 15L,
16L, 17L, 8L, 9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 7L,
8L, 9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 6L, 7L, 8L, 9L,
10L, 11L, 12L, 13L, 14L, 15L, 16L, 5L, 6L, 7L, 8L, 9L, 10L, 11L,
12L, 13L, 14L, 15L, 19L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L,
13L, 14L, 19L, 20L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L,
13L, 18L, 19L, 20L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 17L,
18L, 19L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 15L, 16L, 17L, 18L, 19L,
2L, 3L, 4L, 5L, 6L, 13L, 14L, 15L, 16L, 17L, 18L, 19L, 2L, 3L,
4L, 5L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 18L, 1L, 2L, 3L, 9L,
10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 18L, 1L, 2L, 7L, 8L,
9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 2L, 6L, 7L, 8L, 9L,
10L, 11L, 12L, 13L, 14L, 15L, 16L, 5L, 6L, 7L, 8L, 9L, 10L, 11L,
12L, 13L, 14L, 15L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L,
14L, 4L, 5L, 6L, 7L, 8L, 9L, 10L, 11L, 12L, 13L, 4L, 5L, 6L,
7L, 8L, 9L, 10L, 11L, 6L, 7L, 8L), y = c(1L, 1L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L, 5L, 5L, 5L, 5L, 5L,
5L, 5L, 5L, 5L, 5L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 8L, 8L,
8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 9L, 9L, 9L, 9L,
9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 10L, 10L,
10L, 10L, 10L, 10L, 10L, 10L, 11L, 11L, 11L, 11L, 11L, 11L, 11L,
11L, 11L, 11L, 11L, 11L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L,
12L, 12L, 12L, 12L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L, 13L,
13L, 13L, 13L, 13L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L, 14L,
14L, 14L, 14L, 14L, 15L, 15L, 15L, 15L, 15L, 15L, 15L, 15L, 15L,
15L, 15L, 15L, 16L, 16L, 16L, 16L, 16L, 16L, 16L, 16L, 16L, 16L,
16L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 17L, 18L,
18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 18L, 19L, 19L, 19L, 19L,
19L, 19L, 19L, 19L, 20L, 20L, 20L), col = c("#000000f6", "#000000fd",
"#000000f4", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000f8", "#000000f4", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000fd", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000f9", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000fd",
"#000000f4", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000fa", "#000000ff", "#000000ff", "#000000f6", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000fb", "#000000ff", "#000000ff",
"#000000ff", "#000000f3", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000fa", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000f1", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000f3",
"#000000ff", "#000000ff", "#000000ff", "#000000f6", "#000000f9",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000f5", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000f5",
"#000000fc", "#000000ff", "#000000fd", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000f3", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000ff",
"#000000ff", "#000000f5", "#000000f8", "#000000ff", "#000000ff",
"#000000ff", "#000000ff", "#000000ff", "#000000ff", "#000000f4",
"#000000f1", "#000000fe", "#000000f7"), alpha = c(246L, 253L,
244L, 255L, 255L, 255L, 255L, 255L, 255L, 248L, 244L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 253L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 249L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 253L, 244L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 250L, 255L,
255L, 246L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 251L,
255L, 255L, 255L, 243L, 255L, 255L, 255L, 255L, 255L, 255L, 250L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 241L, 255L,
255L, 255L, 255L, 255L, 243L, 255L, 255L, 255L, 246L, 249L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 245L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 245L, 252L, 255L, 253L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 243L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L,
255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 255L, 245L, 248L,
255L, 255L, 255L, 255L, 255L, 255L, 244L, 241L, 254L, 247L)), row.names = c(NA,
-210L), class = "data.frame")
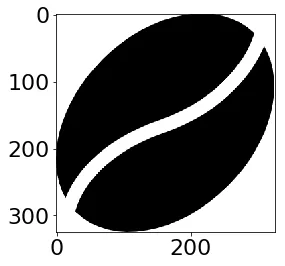
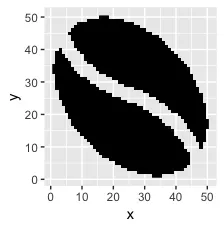
以下是它的样子:
ggplot(sprite_frame, aes(x,y, fill = col)) +
geom_raster() +
guides(fill = F) +
scale_fill_identity()
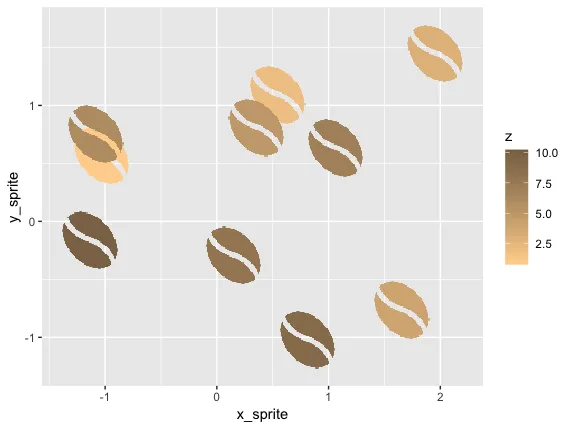
第二步:将这些像素转换为体素
pixels_per_image <- nrow(sprite_frame)
scale <- 1/40
set.seed(2017-02-21)
d <- data.frame(x = rnorm(10), y = rnorm(10), z=1:10)
d2 <- d %>%
mutate(copies = pixels_per_image) %>%
uncount(copies) %>%
mutate(x_sprite = sprite_frame$x*scale + x,
y_sprite = sprite_frame$y*scale + y,
col = rep(sprite_frame$col, nrow(d)))
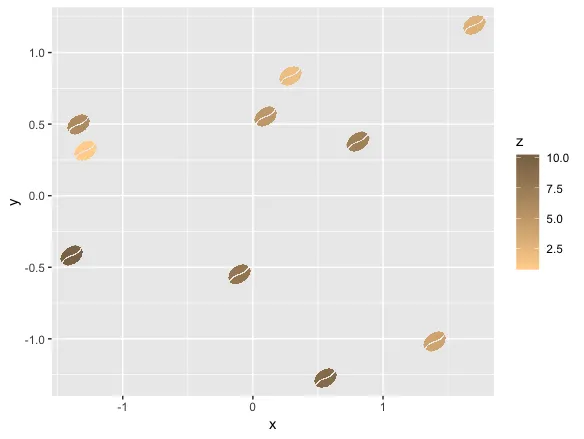
我们可以使用ggplot在二维空间中绘制它:
ggplot(d2, aes(x_sprite, y_sprite, z = z, alpha = col, fill = z)) +
geom_tile(width = scale, height = scale) +
guides(alpha = F) +
scale_fill_gradient(low='burlywood1', high='burlywood4')
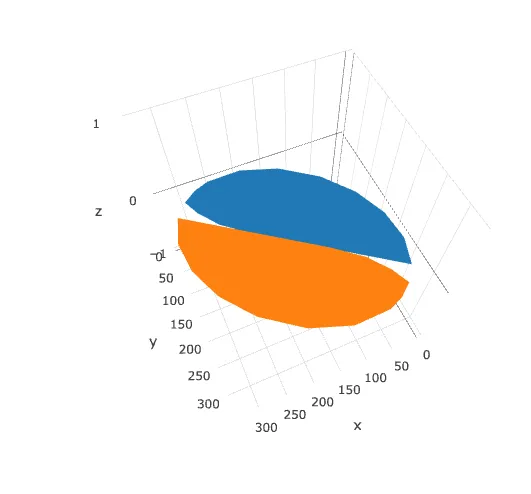
或者将其导入到Plotly中。请注意,目前Plotly 3D散点图不支持可变透明度,因此在缩放到一个精灵时,图像会显示为实心椭圆。
library(plotly)
plot_ly(d2, x = ~x_sprite, y = ~y_sprite, z = ~z,
size = scale, color = ~z, colors = c("#FFD39B", "#8B7355")) %>%
add_markers()
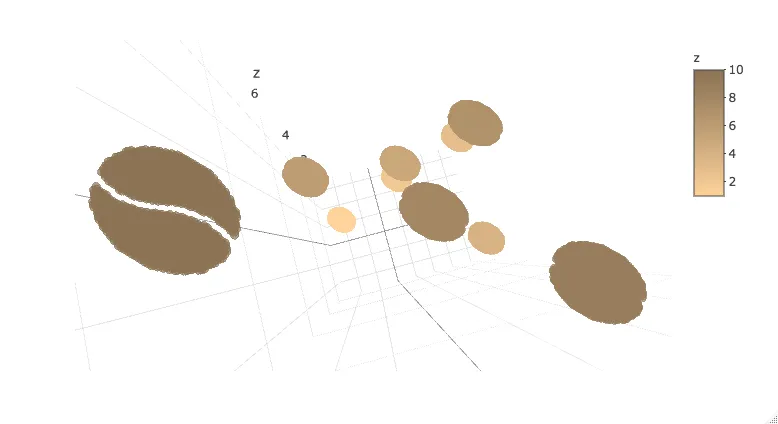
编辑:尝试使用plotly的mesh3d方法
另一种方法似乎是将SVG图形转换为plotly中mesh3d表面的坐标。
我最初尝试这样做的方法非常繁琐:
- 在Inkscape中加载SVG并使用“flatten beziers”选项来近似不带bezier曲线的形状。
- 导出SVG并祈祷文件具有原始坐标。我对SVG还不太熟悉,看起来输出通常可以是绝对和相对点的混合体。在此情况下更加复杂,因为该字形有两个断开的部分。
- 将坐标重新格式化为数据框以便使用ggplot2或plotly进行绘制。
例如,以下坐标表示半个豆子,我们可以通过变换得到另外一半:
library(dplyr)
half_bean <- read.table(
header = T,
stringsAsFactors = F,
text = "x y
153.714 159.412
95.490016 186.286
54.982625 216.85
28.976672 247.7425
14.257 275.602
0.49742188 229.14067
5.610375 175.89737
28.738141 120.85839
69.023 69.01
128.24827 24.564609
190.72412 2.382875
249.14492 3.7247031
274.55165 13.610674
296.205 29.85
296.4 30.064
283.67119 58.138937
258.36 93.03325
216.39731 128.77994
153.714 159.412"
) %>%
mutate(z = 0)
other_half <- half_bean %>%
mutate(x = 330 - x,
y = 330 - y,
z = z)
ggplot() + coord_equal() +
geom_path(data = half_bean, aes(x,y)) +
geom_path(data = other_half, aes(x,y))
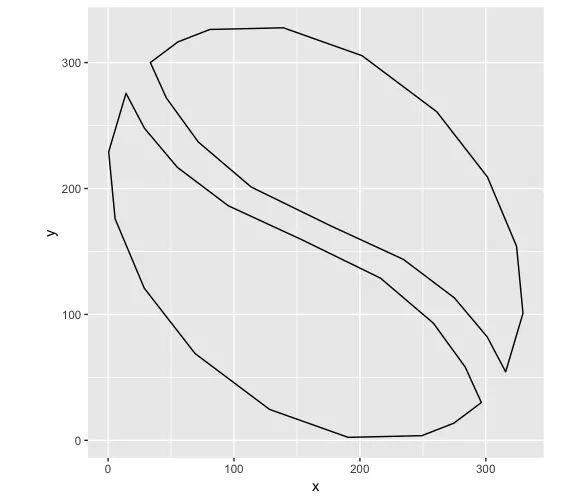
虽然在ggplot中看起来不错,但我在plotly中遇到了显示凹形部分的问题:
library(plotly)
plot_ly(type = 'mesh3d',
split = c(rep(1, 19), rep(2, 19)),
x = c(half_bean$x, other_half$x),
y = c(half_bean$y, other_half$y),
z = c(half_bean$z, other_half$z)
)