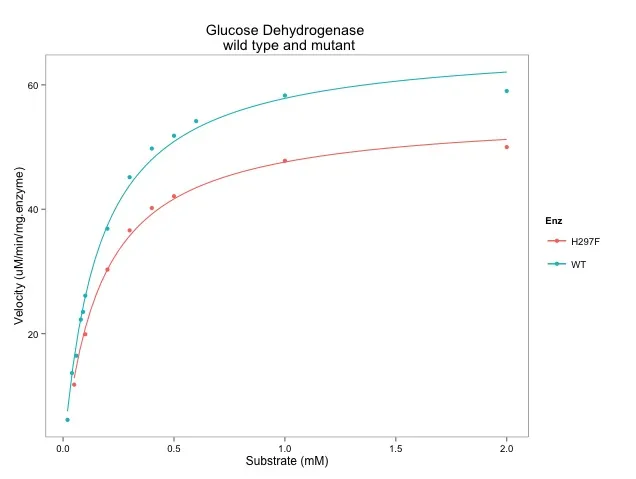
这个this网站发布了有关酶动力学的数据:
Enz <- c("WT","WT","WT","WT","WT",
"WT","WT","WT","WT","WT",
"WT","WT","WT",
"H297F","H297F","H297F",
"H297F","H297F","H297F",
"H297F","H297F")
S <- c(2.00, 1.00, 0.60, 0.50, 0.40,
0.30, 0.20, 0.10, 0.09, 0.08,
0.06, 0.04, 0.02,
0.05, 0.10, 0.20,
0.30, 0.40, 0.50,
1.00, 2.00)
v <- c(59.01, 58.29, 54.17, 51.82, 49.76,
45.15, 36.88, 26.10, 23.50, 22.26,
16.45, 13.67, 6.14,
11.8, 19.9, 30.3,
36.6, 40.2, 42.1,
47.8, 50.0)
绘图的代码:
ggplot(data=enzdata,
aes(x=S,
y=v,
colour = Enz)) +
geom_point() +
xlab("Substrate (mM)") +
ylab("Velocity (uM/min/mg.enzyme)") +
ggtitle("Glucose Dehydrogenase \n wild type and mutant") +
geom_smooth(method = "nls",
formula = y ~ Vmax * x / (Km + x),
start = list(Vmax = 50, Km = 0.2),
se = F, size = 0.5,
data = subset(enzdata, Enz=="WT")) +
geom_smooth(method = "nls",
formula = y ~ Vmax * x / (Km + x),
start = list(Vmax = 50, Km = 0.2),
se = F, size = 0.5,
data = subset(enzdata, Enz=="H297F"))
您是否可以为两条独立曲线添加CI95%带?
使用@adiana的解决方案,它生成了下一个图:
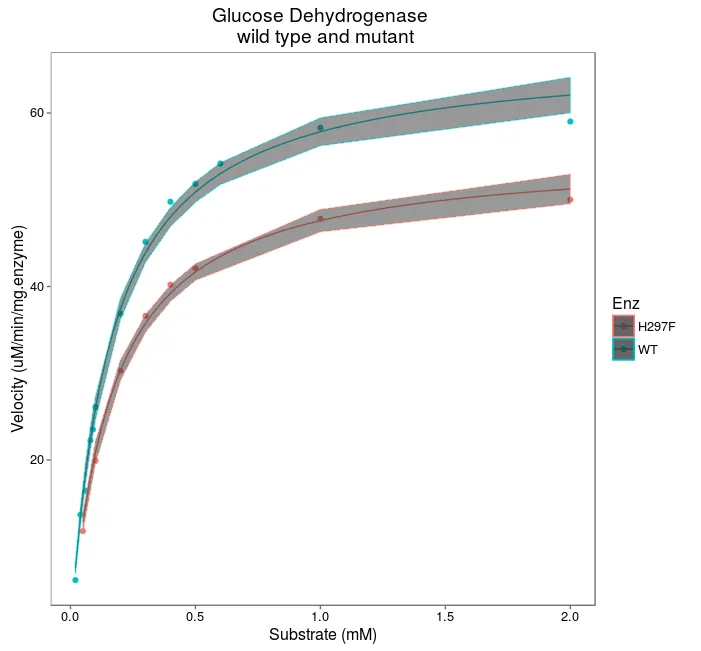
as.lm.nls函数中,我不得不做出以下更改:fo <- as.formula(fo, env = proto:::as.proto.list(L))- c0bra