我有以下的模型:
mod <- glm(data=data, events ~ treatment * size, family = quasipoisson)
以下是输出结果:
Call:
glm(formula = events ~ treatment * size,
family = quasipoisson, data = data)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.4842 -1.4939 -0.4199 0.5921 4.0068
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.070077 0.202376 0.346 0.7299
treatmenttreatment -0.042710 0.315499 -0.135 0.8926
size 0.009464 0.002061 4.591 1.36e-05 ***
treatmenttreatment:size 0.010270 0.005145 1.996 0.0488 *
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for quasipoisson family taken to be 2.039369)
Null deviance: 252.78 on 97 degrees of freedom
Residual deviance: 191.43 on 94 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5
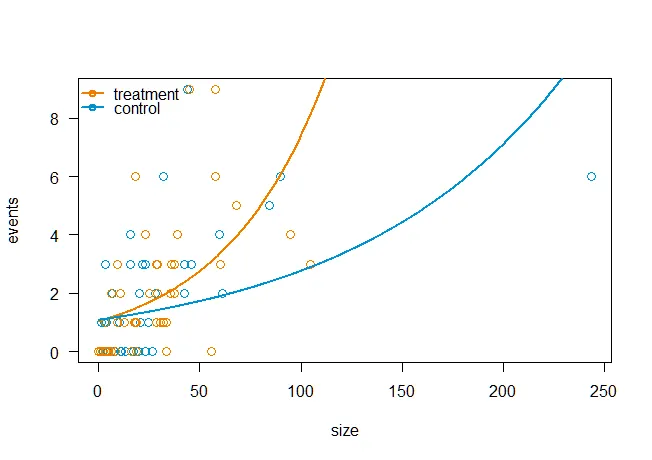
我想使用R基础绘图功能绘制数据图,对于每个组(治疗和对照),使用不同颜色的点,并使用不同颜色的线拟合每个组。类似第20页这里的图表是理想的,但我不知道如何将该示例中使用的Poisson模型转换为我这里的准Poisson模型。下面包含可重复的示例。
individual treatment size events
1 control 10.97 0
2 treatment 0 0
3 control 10.31 1
4 treatment 17.77 1
5 control 11.37 0
6 control 3.857 1
7 treatment 3.8 0
8 treatment 2.029 0
9 treatment 0.8571 0
10 control 0 0
11 control 0 0
12 control 7.943 0
13 control 0 0
14 treatment 8.514 0
15 control 0 0
16 treatment 28.69 1
17 treatment 39.03 4
18 treatment 33.49 0
19 control 2.514 0
20 control 2.771 1
21 treatment 3.257 0
22 control 24.6 1
23 control 1.714 1
24 treatment 9.343 1
25 treatment 10.86 2
26 treatment 28.77 3
27 control 89.97 6
28 control 17.17 0
29 control 4.057 0
30 control 20.4 2
31 treatment 28.49 3
32 treatment 28.66 1
33 treatment 30.66 1
34 control 8.114 0
35 treatment 29.03 2
36 treatment 0 0
37 control 6.543 0
38 treatment 18.86 1
39 control 42.37 3
40 treatment 9.257 3
41 treatment 29 3
42 control 13.46 0
43 control 8.143 0
44 control 0.08571 0
45 treatment 5.2 0
46 control 17.23 0
47 control 17.23 0
48 control 18.97 0
49 treatment 18.4 6
50 treatment 104.6 3
51 control 23.29 3
52 control 3.486 3
53 control 28.2 2
54 control 23 0
55 treatment 37.4 2
56 treatment 16.2 0
57 control 16.03 3
58 treatment 0 0
59 treatment 57.8 6
60 treatment 68.37 5
61 control 4.229 0
62 treatment 45.14 9
63 treatment 33.54 1
64 treatment 55.71 0
65 treatment 12.86 1
66 control 2.429 0
67 treatment 0 0
68 treatment 23.31 4
69 treatment 6.229 2
70 control 21.57 3
71 control 46.11 3
72 treatment 60.29 3
73 control 42.63 2
74 control 61.37 2
75 control 26.8 0
76 treatment 37.57 3
77 treatment 57.83 9
78 control 2.229 0
79 treatment 18.14 1
80 control 19.89 0
81 treatment 35.74 2
82 control 243.6 6
83 control 8.314 0
84 treatment 31.97 1
85 control 84.2 5
86 control 15.91 4
87 treatment 94.66 4
88 treatment 6.429 0
89 treatment 36.2 3
90 control 32.23 6
91 treatment 36.09 3
92 control 43.94 9
93 control 20.86 1
94 control 59.86 4
95 control 7.086 2
96 treatment 3.257 1
97 treatment 18.85 0
98 treatment 25.43 2