我想使用ggplot创建序列图,以保持在我的研究中使用序列分析时相同的视觉风格。我进行以下操作:
library(ggplot2)
library(TraMineR)
library(dplyr)
library(tidyr)
data(mvad)
mvad_seq<-seqdef(mvad,15:length(mvad))
mvad_trate<-seqsubm(mvad_seq,method="TRATE")
mvad_dist<-seqdist(mvad_seq,method="OM",sm=mvad_trate)
cluster<-cutree(hclust(d=as.dist(mvad_dist),method="ward.D2"),k=6)
mvad$cluster<-cluster
mvad_long<-gather(select(mvad,id,contains("."),-matches("N.Eastern"),-matches("S.Eastern")),
key="Month",value="state",
Jul.93, Aug.93, Sep.93, Oct.93, Nov.93, Dec.93, Jan.94, Feb.94, Mar.94,
Apr.94, May.94, Jun.94, Jul.94, Aug.94, Sep.94, Oct.94, Nov.94, Dec.94, Jan.95,
Feb.95, Mar.95, Apr.95, May.95, Jun.95, Jul.95, Aug.95, Sep.95, Oct.95, Nov.95,
Dec.95, Jan.96, Feb.96, Mar.96, Apr.96, May.96, Jun.96, Jul.96, Aug.96, Sep.96,
Oct.96, Nov.96, Dec.96, Jan.97, Feb.97, Mar.97, Apr.97, May.97, Jun.97, Jul.97,
Aug.97, Sep.97, Oct.97, Nov.97, Dec.97, Jan.98, Feb.98, Mar.98, Apr.98, May.98,
Jun.98, Jul.98, Aug.98, Sep.98, Oct.98, Nov.98, Dec.98, Jan.99, Feb.99, Mar.99,
Apr.99, May.99, Jun.99)
mvad_long<-left_join(mvad_long,select(mvad,id,cluster))
ggplot(data=mvad_long,aes(x=Month,y=id,fill=state))+geom_tile()+facet_wrap(~cluster)
我尝试按照聚类绘制序列,结果产生了如下图:
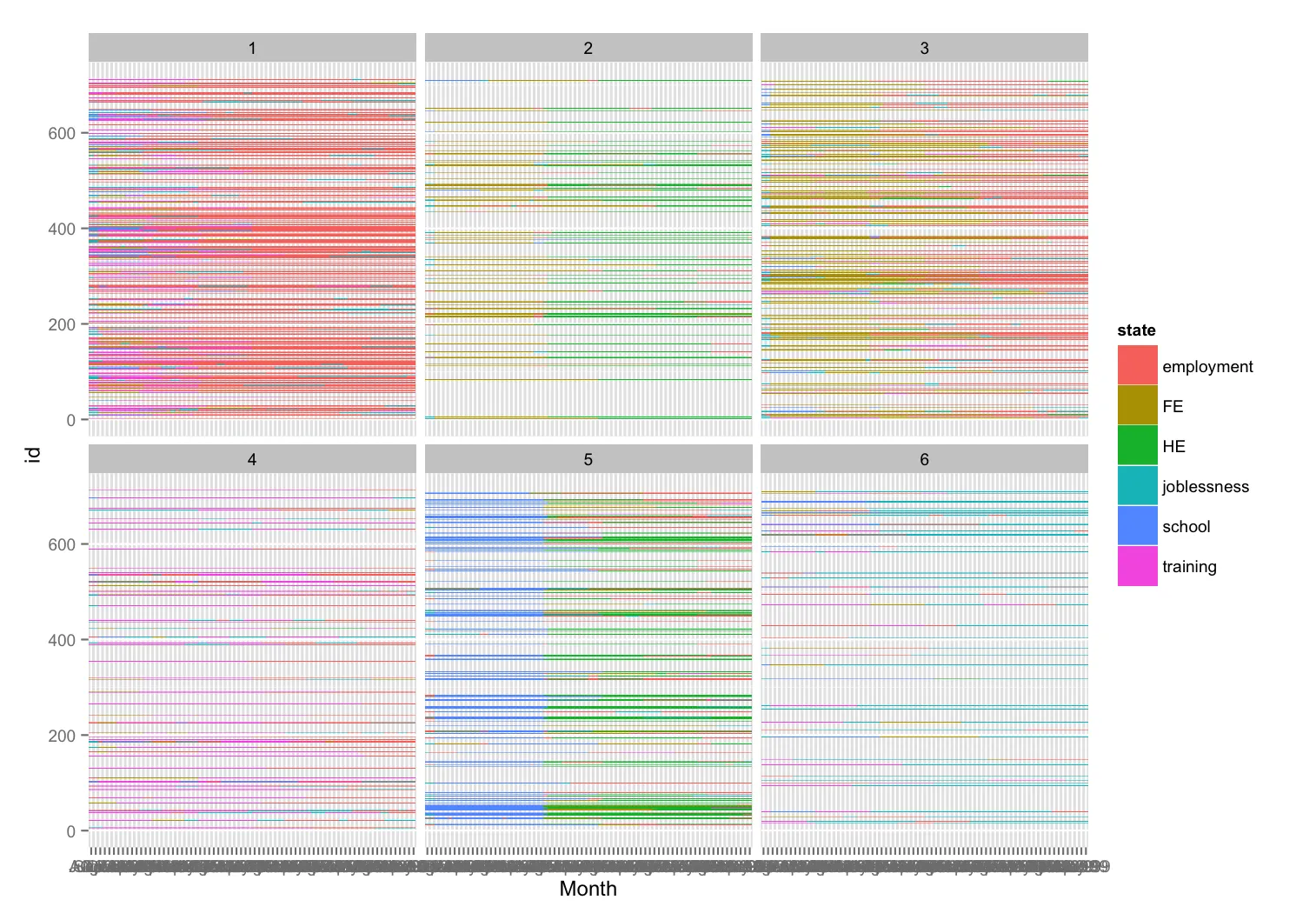 您可以看到,不属于每个面板所代表的聚类的ID存在空缺。我想去掉这些空隙,使得序列像TraMineR的seqIplot()函数一样堆叠显示,结果如下图:
您可以看到,不属于每个面板所代表的聚类的ID存在空缺。我想去掉这些空隙,使得序列像TraMineR的seqIplot()函数一样堆叠显示,结果如下图:
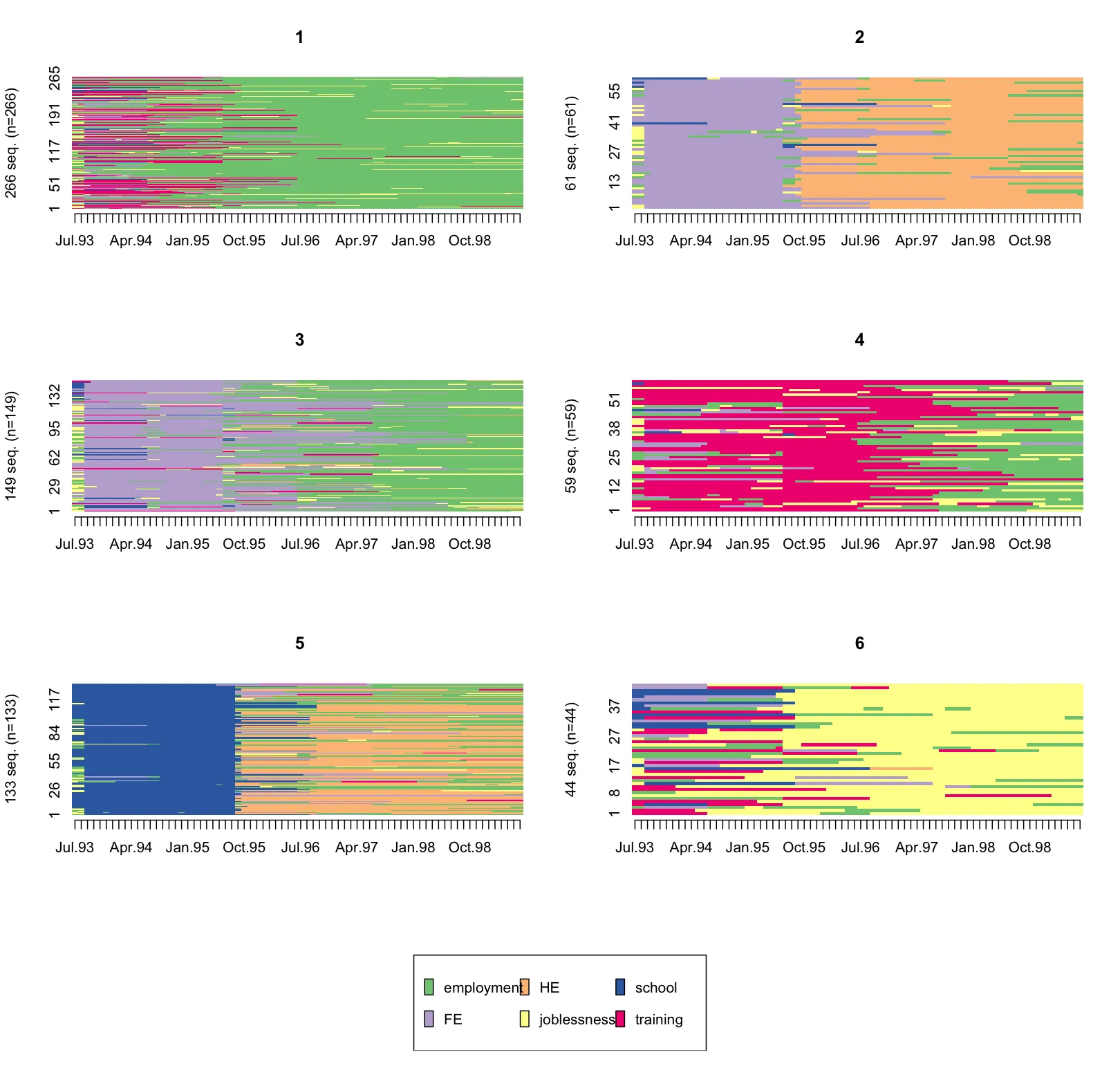 有什么建议吗?
有什么建议吗?
Error in layout_base(data, vars, drop = drop) : At least one layer must contain all variables used for facetting。 - SabDeM