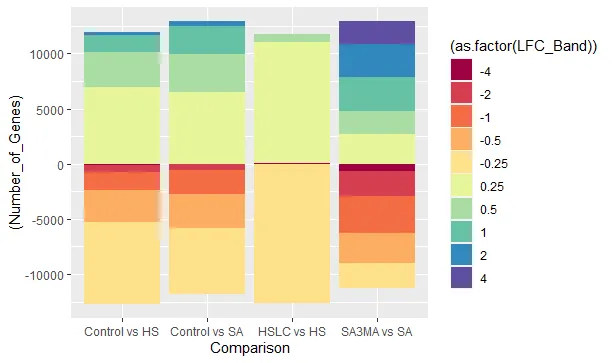
这是我的数据集 -
df<- data.frame(Comparison=c("Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA","Control vs SA",
"SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA","SA3MA vs SA",
"Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS","Control vs HS",
"HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS","HSLC vs HS"),
LFC_Band=c(-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4,
-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4,
-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4,
-4,-2,-1,-0.5,-0.25,0.25,0.5,1,2,4),
Number_of_Genes=c(-36,-540,-2198,-3025,-6002,6517,3462,2466, 500,1,-620,-2317,-3318,-2748,-2264,2706,2046,3079,3013,2073,-119,-606,
-1640,-2886,-7451,6944,3147,1592, 267,0,26,0,0, -44,-901 -11699,10969, 754,0,12,0))
我想要做的是使用ggplot创建一个堆叠条形图。
chart_x <- ggplot(data = df, aes(y=(Number_of_Genes),
x=Comparison, group = Comparison))+
geom_col(aes(fill=(as.factor(LFC_Band))))+
scale_fill_brewer(palette = "Spectral")
chart_x
结果是 - 但是,如果你看到下半部分基本上是翻转的。上半部分是正确的,即从4到0.25,而下半部分则从-4开始。有没有办法翻转下半部分,使其从-0.25开始?简而言之,我想要的是-
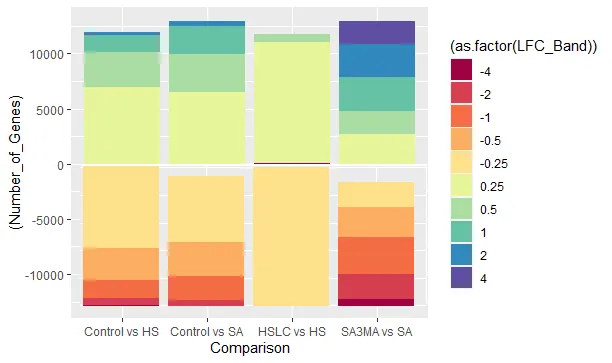 我用MS画图创建了这个图片,只是为了展示我想要的内容。
我用MS画图创建了这个图片,只是为了展示我想要的内容。我尝试过其他方法,例如将数据分成两部分,但效果并不好。任何帮助都将不胜感激。 :)