我已经修改了vioplot函数,使其可以接受列表作为输入,你可以使用这个vioplot2函数:
vioplot2<-function (x, ..., range = 1.5, h = NULL, ylim = NULL, names = NULL,
horizontal = FALSE, col = "magenta", border = "black", lty = 1,
lwd = 1, rectCol = "black", colMed = "white", pchMed = 19,
at, add = FALSE, wex = 1, drawRect = TRUE)
{
if(!is.list(x)){
datas <- list(x, ...)
} else{
datas<-x
}
n <- length(datas)
if (missing(at))
at <- 1:n
upper <- vector(mode = "numeric", length = n)
lower <- vector(mode = "numeric", length = n)
q1 <- vector(mode = "numeric", length = n)
q3 <- vector(mode = "numeric", length = n)
med <- vector(mode = "numeric", length = n)
base <- vector(mode = "list", length = n)
height <- vector(mode = "list", length = n)
baserange <- c(Inf, -Inf)
args <- list(display = "none")
if (!(is.null(h)))
args <- c(args, h = h)
for (i in 1:n) {
data <- datas[[i]]
data.min <- min(data)
data.max <- max(data)
q1[i] <- quantile(data, 0.25)
q3[i] <- quantile(data, 0.75)
med[i] <- median(data)
iqd <- q3[i] - q1[i]
upper[i] <- min(q3[i] + range * iqd, data.max)
lower[i] <- max(q1[i] - range * iqd, data.min)
est.xlim <- c(min(lower[i], data.min), max(upper[i],
data.max))
smout <- do.call("sm.density", c(list(data, xlim = est.xlim),
args))
hscale <- 0.4/max(smout$estimate) * wex
base[[i]] <- smout$eval.points
height[[i]] <- smout$estimate * hscale
t <- range(base[[i]])
baserange[1] <- min(baserange[1], t[1])
baserange[2] <- max(baserange[2], t[2])
}
if (!add) {
xlim <- if (n == 1)
at + c(-0.5, 0.5)
else range(at) + min(diff(at))/2 * c(-1, 1)
if (is.null(ylim)) {
ylim <- baserange
}
}
if (is.null(names)) {
label <- 1:n
}
else {
label <- names
}
boxwidth <- 0.05 * wex
if (!add)
plot.new()
if (!horizontal) {
if (!add) {
plot.window(xlim = xlim, ylim = ylim)
axis(2)
axis(1, at = at, label = label)
}
box()
for (i in 1:n) {
polygon(c(at[i] - height[[i]], rev(at[i] + height[[i]])),
c(base[[i]], rev(base[[i]])), col = col, border = border,
lty = lty, lwd = lwd)
if (drawRect) {
lines(at[c(i, i)], c(lower[i], upper[i]), lwd = lwd,
lty = lty)
rect(at[i] - boxwidth/2, q1[i], at[i] + boxwidth/2,
q3[i], col = rectCol)
points(at[i], med[i], pch = pchMed, col = colMed)
}
}
}
else {
if (!add) {
plot.window(xlim = ylim, ylim = xlim)
axis(1)
axis(2, at = at, label = label)
}
box()
for (i in 1:n) {
polygon(c(base[[i]], rev(base[[i]])), c(at[i] - height[[i]],
rev(at[i] + height[[i]])), col = col, border = border,
lty = lty, lwd = lwd)
if (drawRect) {
lines(c(lower[i], upper[i]), at[c(i, i)], lwd = lwd,
lty = lty)
rect(q1[i], at[i] - boxwidth/2, q3[i], at[i] +
boxwidth/2, col = rectCol)
points(med[i], at[i], pch = pchMed, col = colMed)
}
}
}
invisible(list(upper = upper, lower = lower, median = med,
q1 = q1, q3 = q3))
}
您的示例将得到如下内容:
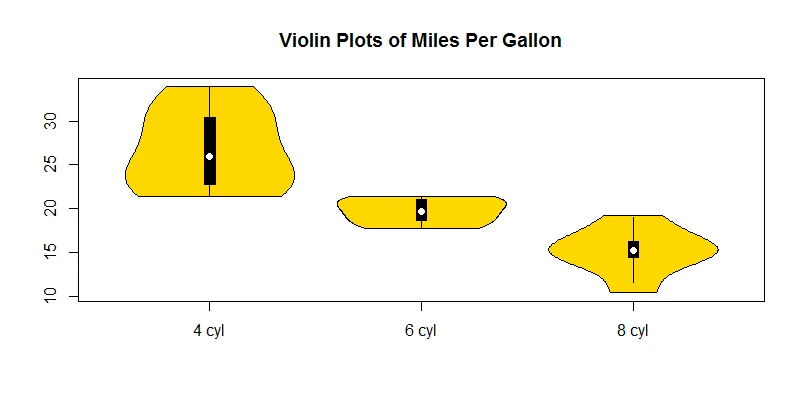

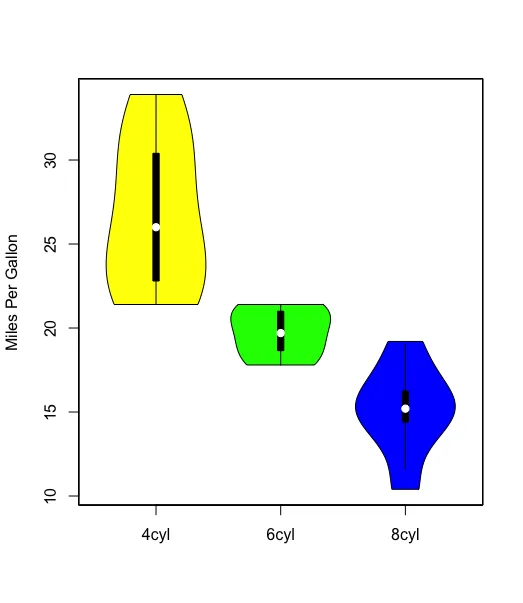
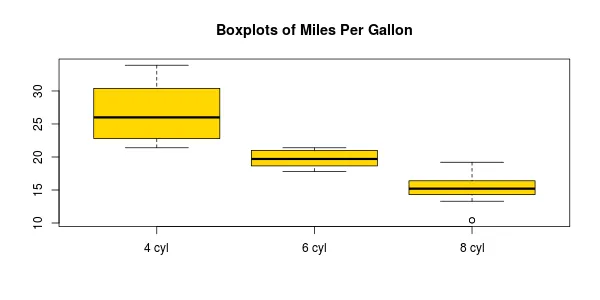
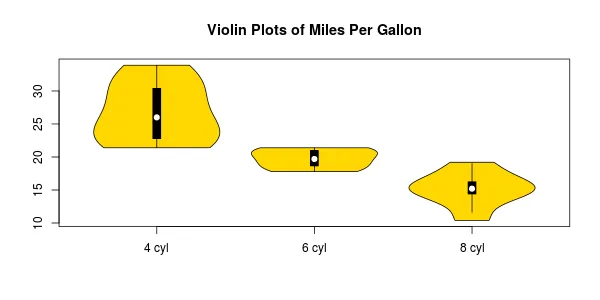
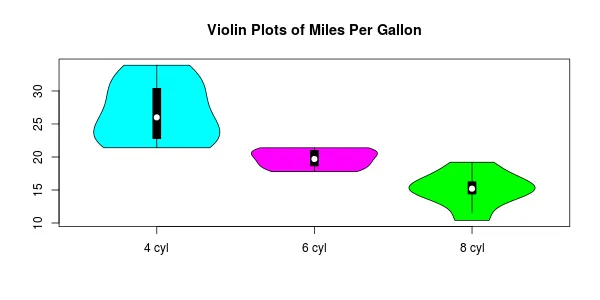
l的更清晰的定义(绕过首先执行所有x的需要)是l <- split(mtcars, mtcars$cyl)。 - Gregor Thomas