我有四个函数用于复杂网络分析中的某些统计计算。
import networkx as nx
import numpy as np
import math
from astropy.io import fits
图的度分布:
def degree_distribution(G):
vk = dict(G.degree())
vk = list(vk.values()) # we get only the degree values
maxk = np.max(vk)
mink = np.min(min)
kvalues= np.arange(0,maxk+1) # possible values of k
Pk = np.zeros(maxk+1) # P(k)
for k in vk:
Pk[k] = Pk[k] + 1
Pk = Pk/sum(Pk) # the sum of the elements of P(k) must to be equal to one
return kvalues,Pk
图的社区检测:
def calculate_community_modularity(graph):
communities = greedy_modularity_communities(graph) # algorithm
modularity_dict = {} # Create a blank dictionary
for i,c in enumerate(communities): # Loop through the list of communities, keeping track of the number for the community
for name in c: # Loop through each neuron in a community
modularity_dict[name] = i # Create an entry in the dictionary for the neuron, where the value is which group they belong to.
nx.set_node_attributes(graph, modularity_dict, 'modularity')
print (graph_name)
for i,c in enumerate(communities): # Loop through the list of communities
#if len(c) > 2: # Filter out modularity classes with 2 or fewer nodes
print('Class '+str(i)+':', len(c)) # Print out the classes and their member numbers
return modularity_dict
图的模块化得分:
def modularity_score(graph):
return nx_comm.modularity(graph, nx_comm.label_propagation_communities(graph))
最后需要展示的是熵图:
def shannon_entropy(G):
k,Pk = degree_distribution(G)
H = 0
for p in Pk:
if(p > 0):
H = H - p*math.log(p, 2)
return H
问题
我现在想要做的是为每个社区(转化为子图)找到本地熵,并保留边缘信息。
这可行吗?怎么做?
编辑
所使用的矩阵可以在此链接中找到:
with fits.open('mind_dataset/matrix_CEREBELLUM_large.fits') as data:
matrix = pd.DataFrame(data[0].data.byteswap().newbyteorder())
然后将邻接矩阵转换成图形,命名为'graph'或者'G':
def matrix_to_graph(matrix):
from_matrix = matrix.copy()
to_numpy = from_matrix.to_numpy()
G = nx.from_numpy_matrix(to_numpy)
return G
编辑2
根据下面提供的答案,我创建了另一个函数:
def community_entropy(modularity_dict):
communities = {}
#create communities as lists of nodes
for node, community in modularity_dict.items():
if community not in communities.keys():
communities[community] = [node]
else:
communities[community].append(node)
print(communities)
#transform lists of nodes to actual subgraphs
for subgraph, community in communities.items():
communities[community] = nx.Graph.subgraph(subgraph)
local_entropy = {}
for subgraph, community in communities.items():
local_entropy[community] = shannon_entropy(subgraph)
return local_entropy
并且:
cerebellum_graph = matrix_to_graph(matrix)
modularity_dict_cereb = calculate_community_modularity(cerebellum_graph)
community_entropy_cereb = community_entropy(modularity_dict_cereb)
但是它会抛出以下错误:
TypeError: subgraph()缺少1个必需的位置参数:'nodes'
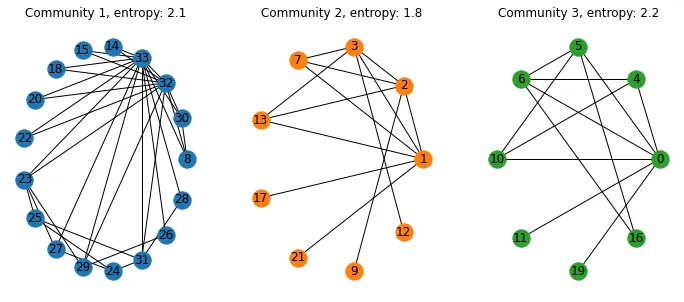
calculate_community_modularity中,您使用greedy_modularity_communities创建一个字典modularity_dict,将图中的节点映射到一个社区。为什么不能将modularity_dict中的每个子图社区传递到shannon_entropy中,以计算该社区的熵? - Warlax56G和graph分别代表什么? - Warlax56