编辑:@Denis 是正确的,一个线性的Rbf(例如 scipy.interpolate.Rbf 并且 "function='linear'") 和 IDW 不同...
(请注意,如果你使用大量点,则所有这些方法都会使用过多的内存!)
以下是IDW的一个简单示例:
def simple_idw(x, y, z, xi, yi):
dist = distance_matrix(x,y, xi,yi)
weights = 1.0 / dist
weights /= weights.sum(axis=0)
zi = np.dot(weights.T, z)
return zi
然而,这里是线性Rbf的定义:
def linear_rbf(x, y, z, xi, yi):
dist = distance_matrix(x,y, xi,yi)
internal_dist = distance_matrix(x,y, x,y)
weights = np.linalg.solve(internal_dist, z)
zi = np.dot(dist.T, weights)
return zi
(在这里使用distance_matrix函数:)
def distance_matrix(x0, y0, x1, y1):
obs = np.vstack((x0, y0)).T
interp = np.vstack((x1, y1)).T
d0 = np.subtract.outer(obs[:,0], interp[:,0])
d1 = np.subtract.outer(obs[:,1], interp[:,1])
return np.hypot(d0, d1)
将所有内容组合成一个漂亮的复制粘贴示例,可以得到一些快速比较图:
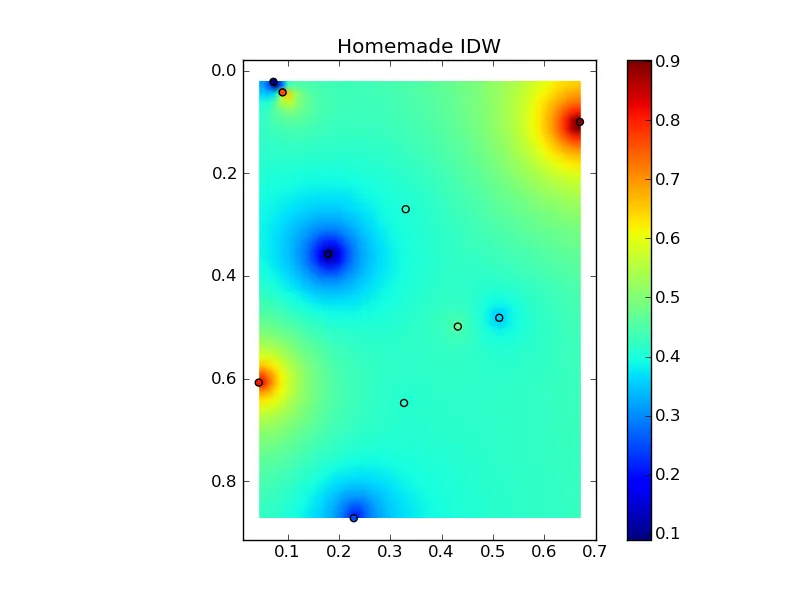 (来源:www.geology.wisc.edu上的jkington)
(来源:www.geology.wisc.edu上的jkington)
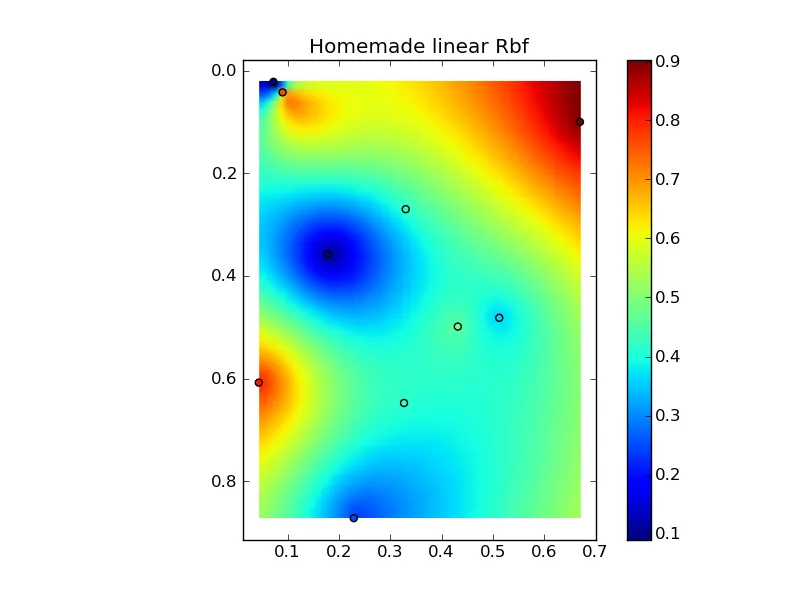 (来源:www.geology.wisc.edu上的jkington)
(来源:www.geology.wisc.edu上的jkington)
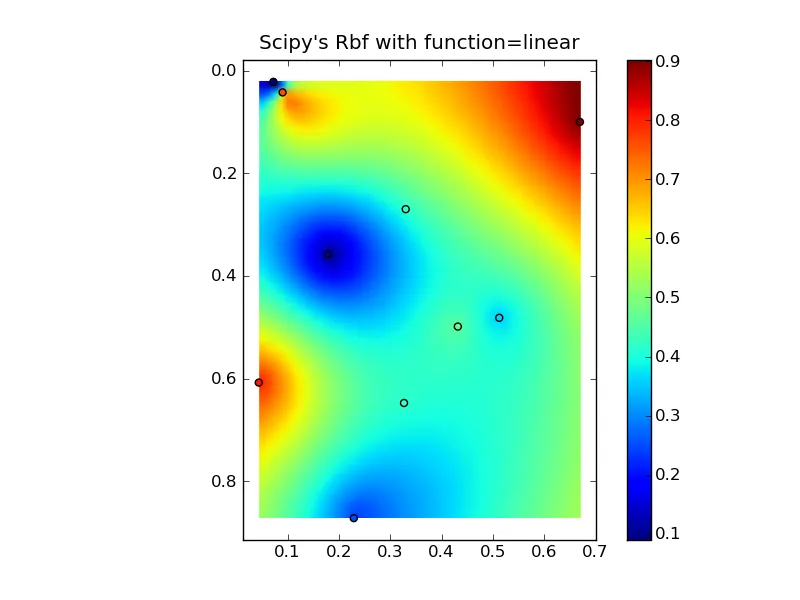 (来源:www.geology.wisc.edu上的jkington)
(来源:www.geology.wisc.edu上的jkington)
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import Rbf
def main():
n = 10
nx, ny = 50, 50
x, y, z = map(np.random.random, [n, n, n])
xi = np.linspace(x.min(), x.max(), nx)
yi = np.linspace(y.min(), y.max(), ny)
xi, yi = np.meshgrid(xi, yi)
xi, yi = xi.flatten(), yi.flatten()
grid1 = simple_idw(x,y,z,xi,yi)
grid1 = grid1.reshape((ny, nx))
grid2 = scipy_idw(x,y,z,xi,yi)
grid2 = grid2.reshape((ny, nx))
grid3 = linear_rbf(x,y,z,xi,yi)
print grid3.shape
grid3 = grid3.reshape((ny, nx))
plot(x,y,z,grid1)
plt.title('Homemade IDW')
plot(x,y,z,grid2)
plt.title("Scipy's Rbf with function=linear")
plot(x,y,z,grid3)
plt.title('Homemade linear Rbf')
plt.show()
def simple_idw(x, y, z, xi, yi):
dist = distance_matrix(x,y, xi,yi)
weights = 1.0 / dist
weights /= weights.sum(axis=0)
zi = np.dot(weights.T, z)
return zi
def linear_rbf(x, y, z, xi, yi):
dist = distance_matrix(x,y, xi,yi)
internal_dist = distance_matrix(x,y, x,y)
weights = np.linalg.solve(internal_dist, z)
zi = np.dot(dist.T, weights)
return zi
def scipy_idw(x, y, z, xi, yi):
interp = Rbf(x, y, z, function='linear')
return interp(xi, yi)
def distance_matrix(x0, y0, x1, y1):
obs = np.vstack((x0, y0)).T
interp = np.vstack((x1, y1)).T
d0 = np.subtract.outer(obs[:,0], interp[:,0])
d1 = np.subtract.outer(obs[:,1], interp[:,1])
return np.hypot(d0, d1)
def plot(x,y,z,grid):
plt.figure()
plt.imshow(grid, extent=(x.min(), x.max(), y.max(), y.min()))
plt.hold(True)
plt.scatter(x,y,c=z)
plt.colorbar()
if __name__ == '__main__':
main()


