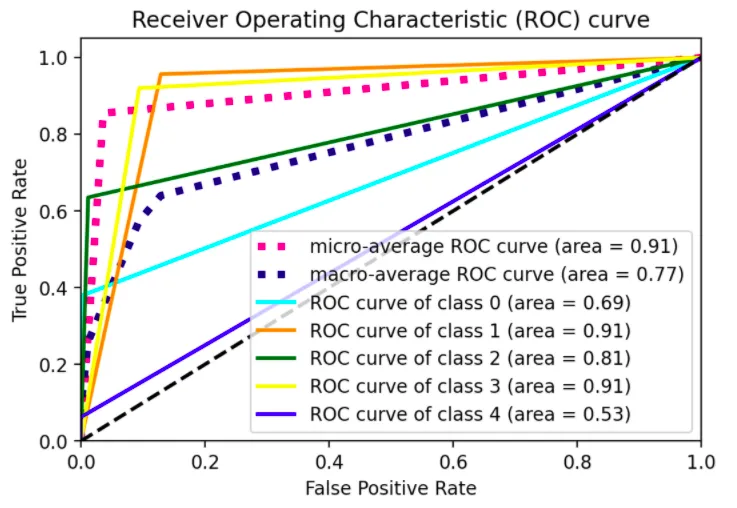
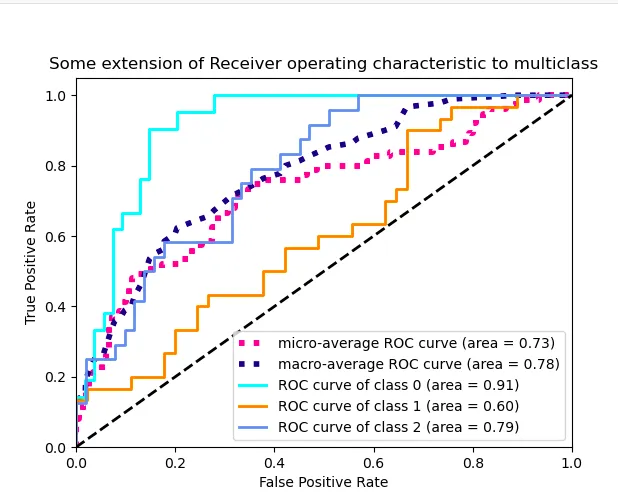
sklearn的 ROC扩展到多类的思想应用于我的数据集。我的每个类别的ROC曲线看起来都像是一条直线,与sklearn示例中波动的曲线不同。下面是我提供的一个MWE以说明我的意思:
# all imports
import numpy as np
import matplotlib.pyplot as plt
from itertools import cycle
from sklearn import svm, datasets
from sklearn.metrics import roc_curve, auc
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import label_binarize
from sklearn.datasets import make_classification
from sklearn.ensemble import RandomForestClassifier
# dummy dataset
X, y = make_classification(10000, n_classes=5, n_informative=10, weights=[.04, .4, .12, .5, .04])
train, test, ytrain, ytest = train_test_split(X, y, test_size=.3, random_state=42)
# random forest model
model = RandomForestClassifier()
model.fit(train, ytrain)
yhat = model.predict(test)
以下函数绘制ROC曲线:
def plot_roc_curve(y_test, y_pred):
n_classes = len(np.unique(y_test))
y_test = label_binarize(y_test, classes=np.arange(n_classes))
y_pred = label_binarize(y_pred, classes=np.arange(n_classes))
# Compute ROC curve and ROC area for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], y_pred[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Compute micro-average ROC curve and ROC area
fpr["micro"], tpr["micro"], _ = roc_curve(y_test.ravel(), y_pred.ravel())
roc_auc["micro"] = auc(fpr["micro"], tpr["micro"])
# First aggregate all false positive rates
all_fpr = np.unique(np.concatenate([fpr[i] for i in range(n_classes)]))
# Then interpolate all ROC curves at this points
mean_tpr = np.zeros_like(all_fpr)
for i in range(n_classes):
mean_tpr += np.interp(all_fpr, fpr[i], tpr[i])
# Finally average it and compute AUC
mean_tpr /= n_classes
fpr["macro"] = all_fpr
tpr["macro"] = mean_tpr
roc_auc["macro"] = auc(fpr["macro"], tpr["macro"])
# Plot all ROC curves
#plt.figure(figsize=(10,5))
plt.figure(dpi=600)
lw = 2
plt.plot(fpr["micro"], tpr["micro"],
label="micro-average ROC curve (area = {0:0.2f})".format(roc_auc["micro"]),
color="deeppink", linestyle=":", linewidth=4,)
plt.plot(fpr["macro"], tpr["macro"],
label="macro-average ROC curve (area = {0:0.2f})".format(roc_auc["macro"]),
color="navy", linestyle=":", linewidth=4,)
colors = cycle(["aqua", "darkorange", "darkgreen", "yellow", "blue"])
for i, color in zip(range(n_classes), colors):
plt.plot(fpr[i], tpr[i], color=color, lw=lw,
label="ROC curve of class {0} (area = {1:0.2f})".format(i, roc_auc[i]),)
plt.plot([0, 1], [0, 1], "k--", lw=lw)
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic (ROC) curve")
plt.legend()
输出:
plot_roc_curve(ytest, yhat)
一条直线弯曲一次。我希望能够看到不同阈值下的模型表现,而不仅仅是一个阈值,类似于sklearn的示例中展示的3类情况的图形: