我的图表中的图例被裁剪了。为了解决这个问题,我知道我需要处理边距,但是我不知道该如何操作。而且似乎使用xpd=TRUE也无效。
我的代码结构如下:
plot(x,y1)
par(new=TRUE)
plot(x,y2)
par(new=TRUE)
plot(x,y3)
...
par(xpd=TRUE)
legend(...)
整个代码:
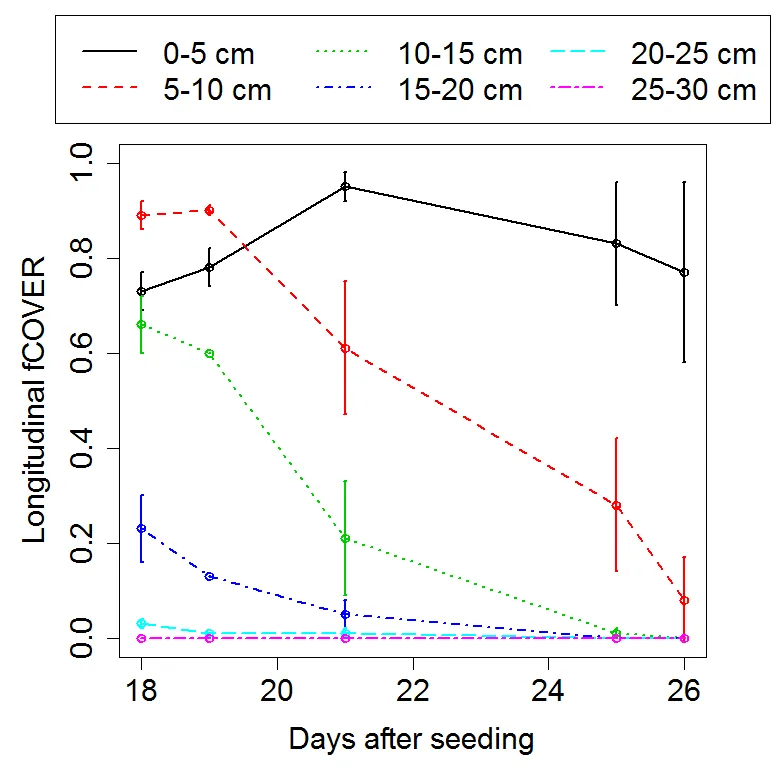
time<-c(18,19, 21, 25, 26)
layer_0<-c(0.73,0.78,0.95,0.83,0.77)
layer_0_sd<-c(0.04,0.04,0.03,0.13,0.19)
layer_1<-c(0.89,0.9,0.61,0.28,0.08)
layer_1_sd<-c(0.03,0.01,0.14,0.14,0.09)
layer_2<-c(0.66,0.6,0.21,0.01,0)
layer_2_sd<-c(0.06,0,0.12,0.01,0)
layer_3<-c(0.23,0.13,0.05,0,0)
layer_3_sd<-c(0.07,0,0.03,0,0)
layer_4<-c(0.03,0.01,0.01,0,0)
layer_4_sd<-c(0.01,0,0.01,0,0)
layer_5<-c(0,0,0,0,0)
layer_5_sd<-c(0,0,0,0,0)
epsilon=0.02
plot(time, layer_0, ylim=c(0,1), type="o", lty=1, lwd=2,ylab="Longitudinal fCOVER", xlab="Days after seeding", cex.lab=1.5, cex.axis=1.5, cex.main=1.5, cex.sub=1.5)
segments(time, layer_0-layer_0_sd,time, layer_0+layer_0_sd, lwd=2)
segments(time-epsilon,layer_0-layer_0_sd,time+epsilon,layer_0-layer_0_sd, lwd=2)
segments(time-epsilon,layer_0+layer_0_sd,time+epsilon,layer_0+layer_0_sd, lwd=2)
for (i in c(1:5)){
par(new=TRUE)
eval(parse(text=paste("plot(time, layer_",i,", ylim=c(0,1), type='o', xlab='', ylab='', xaxt='n', yaxt='n',lwd=2, lty=",i+1,", col=",i+1,")+
segments(time, layer_",i,"-layer_",i,"_sd,time, layer_",i,"+layer_",i,"_sd, col=",i+1,",lwd=2)+
segments(time-epsilon,layer_",i,"-layer_",i,"_sd,time+epsilon,layer_",i,"-layer_",i,"_sd, col=",i+1,",lwd=2)+
segments(time-epsilon,layer_",i,"+layer_",i,"_sd,time+epsilon,layer_",i,"+layer_",i,"_sd, col=",i+1,",lwd=2)", sep="")))
}
par(xpd=TRUE)
legend(x=17.65, y=1.3, c("0-5 cm","5-10 cm", "10-15 cm", "15-20 cm", "20-25 cm","25-30 cm"), lty=c(1,2,3,4,5,6), col=c(1,2,3,4,5,6),ncol=3, lwd=2, cex=1.5)
?par吗?请确保你的代码可复现,这样我们回答起来会更容易。 - Roland?legend可能会更有帮助,特别是关于“inset”参数的部分。 - IRTFM