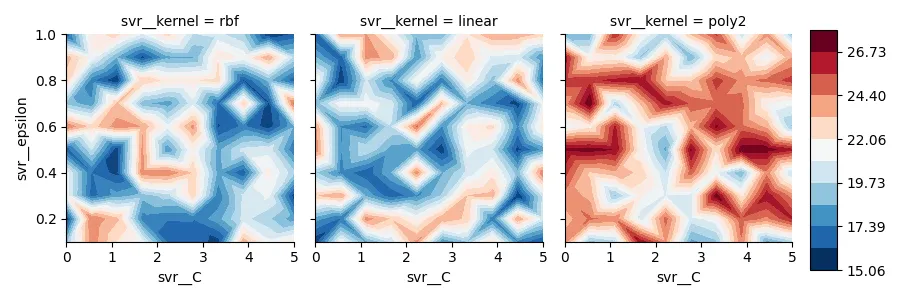
为了找到最佳的超参数来支持向量回归,我进行了网格搜索,并获得了一个类似于DataFrame的结果。
为了可视化结果,我为一个核函数创建了等高线图。
svr__kernel svr__C svr__epsilon mae
rbf 0.01 0.1 19.80
linear 0.01 0.1 19.00
poly2 0.01 0.1 19.72
rbf 0.01 0.2 19.76
.. .. .. ..
为了可视化结果,我为一个核函数创建了等高线图。
fig, ax = plt.subplots(figsize=(15,7))
plot_df = df[df.svr__kernel == "poly2"].copy()
C = plot_df["svr__C"]
epsilon = plot_df["svr__epsilon"]
score = plot_df["mae"]
# Plotting all evaluations:
ax.plot(C, epsilon, "ko", ms=1)
# Create a contour plot
cntr = ax.tricontourf(C, epsilon, score, levels=12, cmap="RdBu_r")
# Adjusting the colorbar
fig.colorbar(cntr, ax=ax, label="MAE")
# Adjusting the axis limits
ax.set(
xlim=(min(C), max(C)),
ylim=(min(epsilon), max(epsilon)),
xlabel="C",
ylabel="Epsilon",
)
ax.set_title("SVR performance landscape")
现在我想用每个核的轮廓图来创建FacetGrid,并使用相同的颜色条对mae值进行比较。不幸的是,我对FacetGrid的操作流程存在严重问题。