一种快速的numpy方法是填充数组(如果您想考虑边缘),然后比较移动的切片。请参见下面的localMax函数。
import numpy as np
import matplotlib.pyplot as plt
Nx,Ny = 100,200
grid = np.mgrid[-1:1:1j*Nx,-1:1:1j*Ny]
filt = 10**(-10*(1-grid**2).mean(axis=0))
cloudy = np.abs(np.fft.ifft2((np.random.rand(Nx,Ny)-.5)*filt))/filt.mean()
Nx,Ny = 10,20
checkerboard = 1.0*np.mgrid[0:Nx,0:Ny].sum(axis=0)%2
checkerboard *= 2-(np.mgrid[-1:1:1j*Nx,-1:1:1j*Ny]**2).sum(axis=0)
def localMax(a, include_diagonal=True, threshold=-np.inf) :
ap = np.pad(a, ((1,1),(1,1)), constant_values=-np.inf )
adjacentmax =(
(ap[1:-1,1:-1] > threshold) &
(ap[0:-2,1:-1] <= ap[1:-1,1:-1]) &
(ap[2:, 1:-1] <= ap[1:-1,1:-1]) &
(ap[1:-1,0:-2] <= ap[1:-1,1:-1]) &
(ap[1:-1,2: ] <= ap[1:-1,1:-1])
)
if not include_diagonal :
return np.argwhere(adjacentmax)
diagonalmax =(
(ap[0:-2,0:-2] <= ap[1:-1,1:-1]) &
(ap[2: ,2: ] <= ap[1:-1,1:-1]) &
(ap[0:-2,2: ] <= ap[1:-1,1:-1]) &
(ap[2: ,0:-2] <= ap[1:-1,1:-1])
)
return np.argwhere(adjacentmax & diagonalmax)
plt.figure(1); plt.clf()
plt.imshow(cloudy, cmap='bone')
mx1 = localMax(cloudy)
mx2 = localMax(cloudy, threshold=cloudy.mean()*.8)
plt.scatter(mx1[:,1],mx1[:,0], color='yellow', s=20)
plt.scatter(mx2[:,1],mx2[:,0], color='red', s=5)
plt.savefig('localMax1.png')
plt.figure(2); plt.clf()
plt.imshow(checkerboard, cmap='bone')
mx1 = localMax(checkerboard,False)
mx2 = localMax(checkerboard)
plt.scatter(mx1[:,1],mx1[:,0], color='yellow', s=20)
plt.scatter(mx2[:,1],mx2[:,0], color='red', s=10)
plt.savefig('localMax2.png')
plt.show()
时间与scipy过滤器大致相同:
In [169]: %timeit mx2 = np.argwhere((maximum_filter(cloudy, size=3)==cloudy) & (cloudy>.5))
244 µs ± 1.1 µs per loop (mean ± std. dev. of 7 runs, 1000 loops each)
In [172]: %timeit mx1 = localMax(cloudy, True, .5)
每个循环262微秒±1.44微秒(平均值±7次运行的标准差,每个1000次循环)
这是带有所有峰值(黄色)和高于阈值峰值(红色)的“多云”示例:
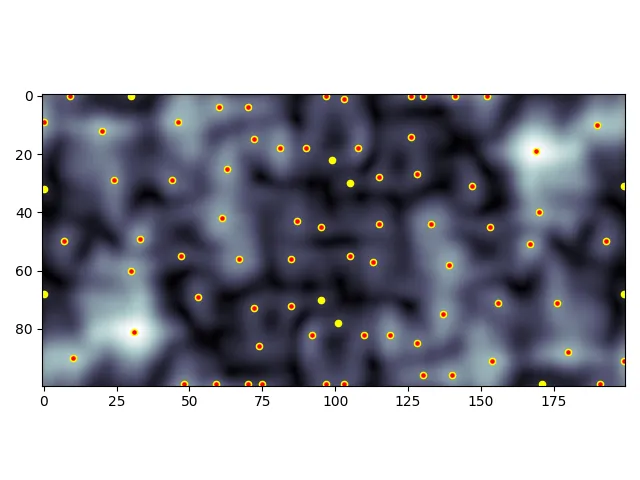
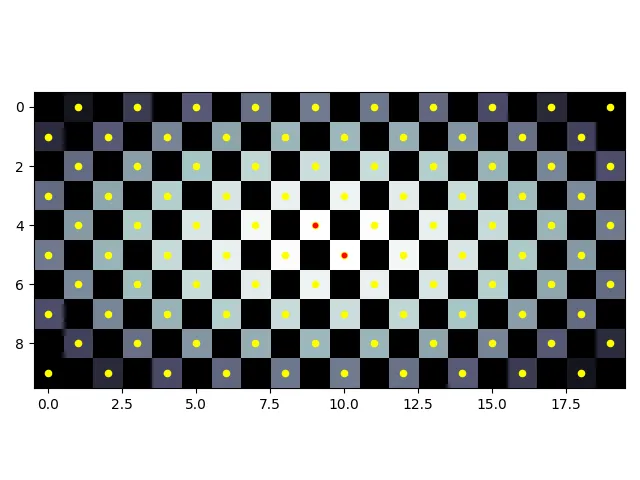
根据您的用例是否包括对角线可能很重要。棋盘演示了不考虑对角线的峰值(黄色)和考虑对角线的峰值(红色):
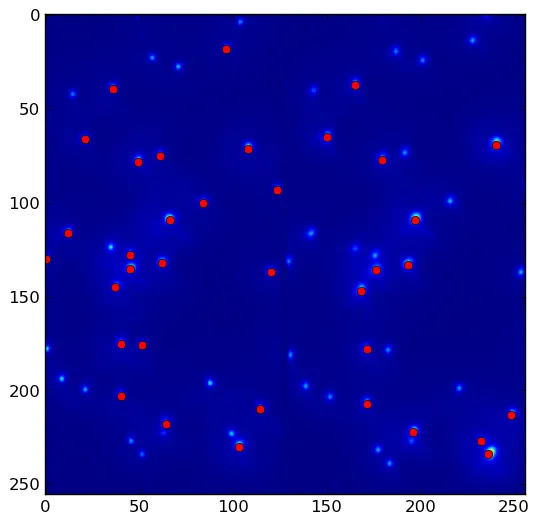


(x,y)坐标的方法吗?如果是这样,你可以使用np.where(maxima)找到它们。 - unutbu