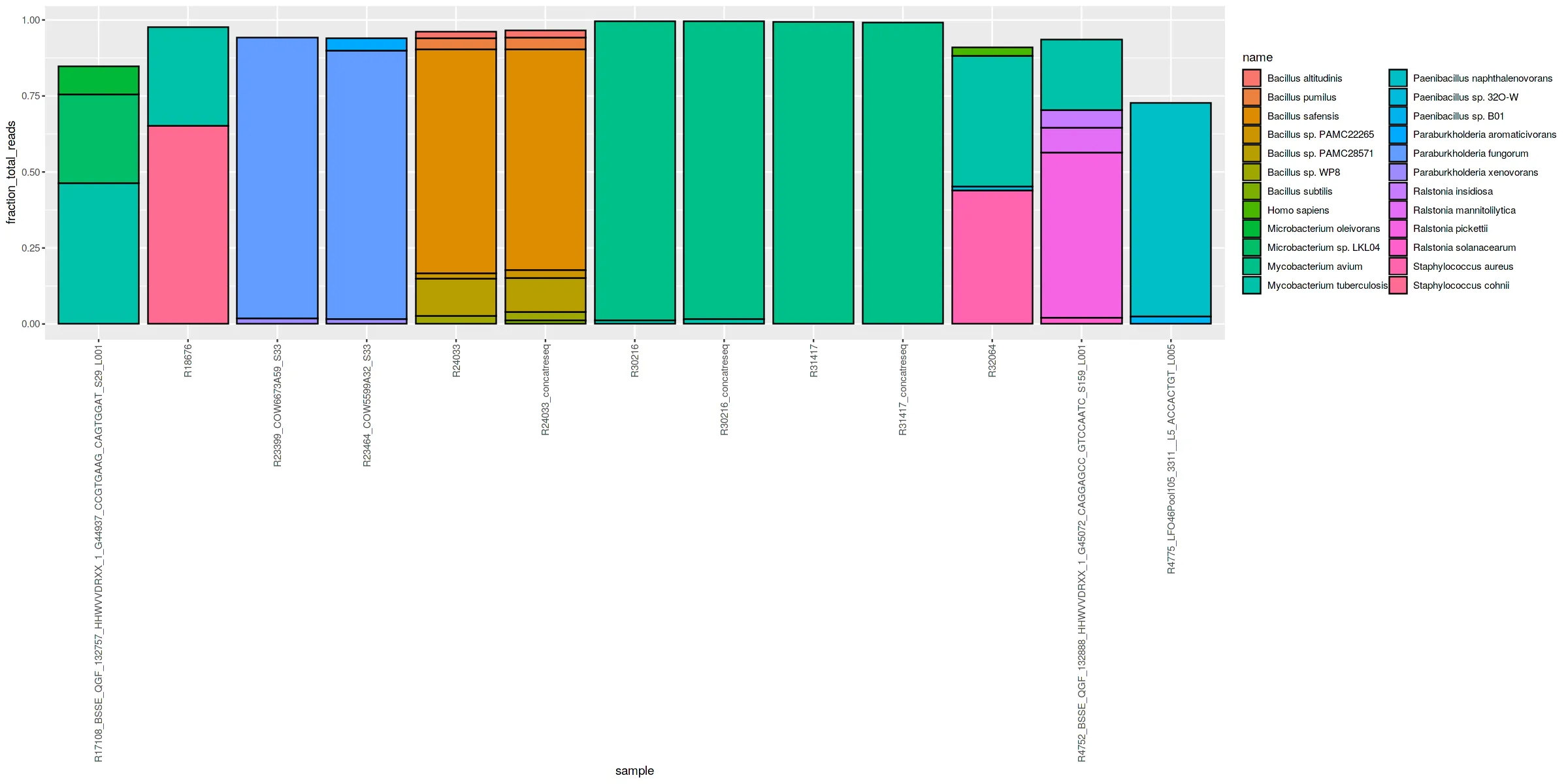
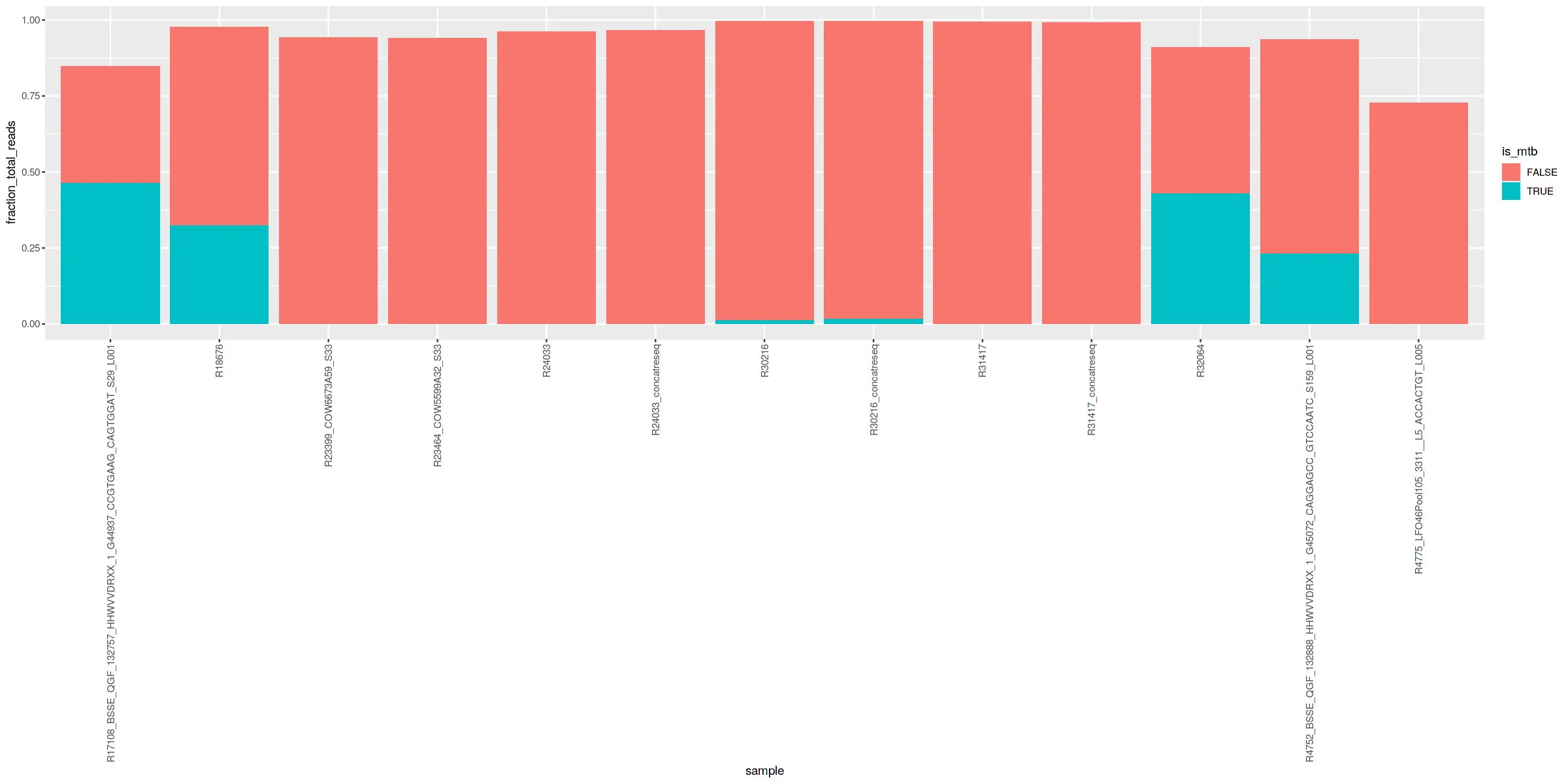
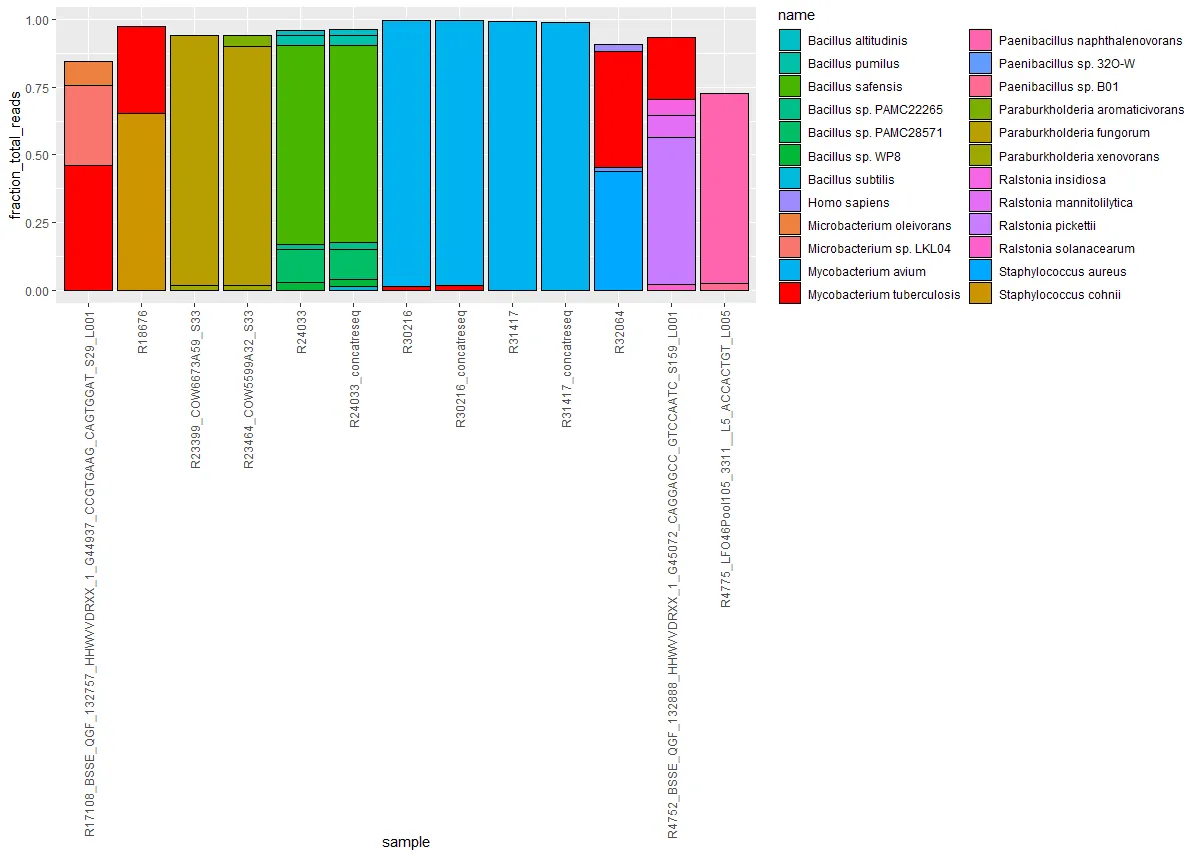
我有几个实验的数据,这些数据给出了样本中要表示的物种DNA比例,我想用堆积条形图来表示。以下是示例数据:
sample_df <- structure(list(sample = c("R17108_BSSE_QGF_132757_HHWVVDRXX_1_G44937_CCGTGAAG_CAGTGGAT_S29_L001",
"R17108_BSSE_QGF_132757_HHWVVDRXX_1_G44937_CCGTGAAG_CAGTGGAT_S29_L001",
"R17108_BSSE_QGF_132757_HHWVVDRXX_1_G44937_CCGTGAAG_CAGTGGAT_S29_L001",
"R18676", "R18676", "R23399_COW6673A59_S33", "R23399_COW6673A59_S33",
"R23464_COW5599A32_S33", "R23464_COW5599A32_S33", "R23464_COW5599A32_S33",
"R24033_concatreseq", "R24033_concatreseq", "R24033_concatreseq",
"R24033_concatreseq", "R24033_concatreseq", "R24033_concatreseq",
"R24033_concatreseq", "R24033", "R24033", "R24033", "R24033",
"R24033", "R24033", "R30216_concatreseq", "R30216_concatreseq",
"R30216", "R30216", "R31417_concatreseq", "R31417", "R32064",
"R32064", "R32064", "R32064", "R4752_BSSE_QGF_132888_HHWVVDRXX_1_G45072_CAGGAGCC_GTCCAATC_S159_L001",
"R4752_BSSE_QGF_132888_HHWVVDRXX_1_G45072_CAGGAGCC_GTCCAATC_S159_L001",
"R4752_BSSE_QGF_132888_HHWVVDRXX_1_G45072_CAGGAGCC_GTCCAATC_S159_L001",
"R4752_BSSE_QGF_132888_HHWVVDRXX_1_G45072_CAGGAGCC_GTCCAATC_S159_L001",
"R4752_BSSE_QGF_132888_HHWVVDRXX_1_G45072_CAGGAGCC_GTCCAATC_S159_L001",
"R4775_LFO46Pool105_3311__L5_ACCACTGT_L005", "R4775_LFO46Pool105_3311__L5_ACCACTGT_L005"
), name = c("Microbacterium sp. LKL04", "Microbacterium oleivorans",
"Mycobacterium tuberculosis", "Staphylococcus cohnii", "Mycobacterium tuberculosis",
"Paraburkholderia fungorum", "Paraburkholderia xenovorans", "Paraburkholderia fungorum",
"Paraburkholderia aromaticivorans", "Paraburkholderia xenovorans",
"Bacillus safensis", "Bacillus sp. WP8", "Bacillus sp. PAMC28571",
"Bacillus sp. PAMC22265", "Bacillus pumilus", "Bacillus altitudinis",
"Bacillus subtilis", "Bacillus safensis", "Bacillus sp. WP8",
"Bacillus sp. PAMC28571", "Bacillus sp. PAMC22265", "Bacillus pumilus",
"Bacillus altitudinis", "Mycobacterium avium", "Mycobacterium tuberculosis",
"Mycobacterium avium", "Mycobacterium tuberculosis", "Mycobacterium avium",
"Mycobacterium avium", "Staphylococcus aureus", "Paenibacillus sp. 32O-W",
"Mycobacterium tuberculosis", "Homo sapiens", "Ralstonia pickettii",
"Ralstonia mannitolilytica", "Ralstonia insidiosa", "Ralstonia solanacearum",
"Mycobacterium tuberculosis", "Paenibacillus naphthalenovorans",
"Paenibacillus sp. B01"), fraction_total_reads = c(0.29347, 0.09071,
0.46242, 0.6525, 0.32403, 0.92541, 0.01772, 0.8842, 0.04011,
0.01561, 0.72733, 0.02744, 0.11121, 0.02673, 0.03845, 0.02282,
0.01176, 0.73674, 0.02711, 0.12122, 0.01858, 0.03677, 0.02115,
0.97964, 0.01579, 0.98397, 0.01227, 0.9907, 0.99348, 0.43967,
0.01337, 0.4288, 0.02825, 0.54439, 0.08077, 0.05916, 0.01978,
0.23135, 0.70247, 0.02424)), row.names = c(NA, -40L), class = c("tbl_df",
"tbl", "data.frame"))
然后像这样绘制它
ggplot(data = sample_df, aes(fill=name, y=fraction_total_reads, x=sample)) +
geom_bar(position="stack", stat="identity", color="black") +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
问题是我主要关心结核分枝杆菌,但它的颜色经常在其他分枝杆菌物种中丢失,因此不突出显示。是否有一种方法可以为一个物种分配特定的不同颜色、背景或边框,而不必为所有物种分配颜色?我的真实数据有1000多个物种,这将非常费力。在其他地方,我看到建议像以下方式创建新列
sample_df2 <- sample_df %>%
mutate(is_mtb = ifelse(name == "Mycobacterium tuberculosis", TRUE, FALSE))
然后使用plot进行绘图。
ggplot(data = sample_df2, aes(fill=is_mtb, y=fraction_total_reads, x=sample)) +
geom_bar(position="stack", stat="identity") +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1))
如果有一个像第一个那样突出显示结核分枝杆菌,并且在每个条形图的底部标注,对我来说会更好。