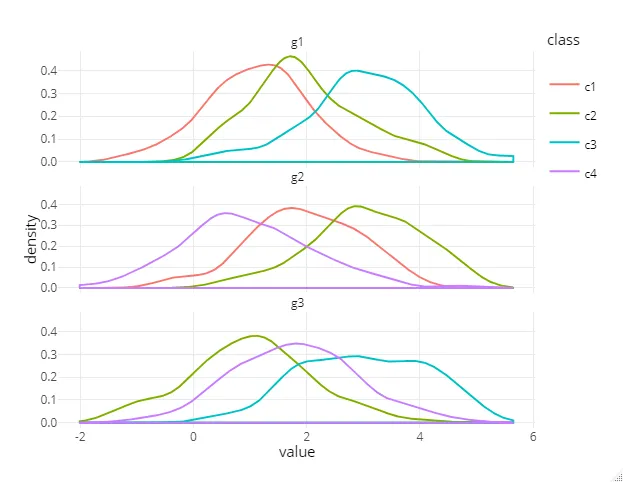
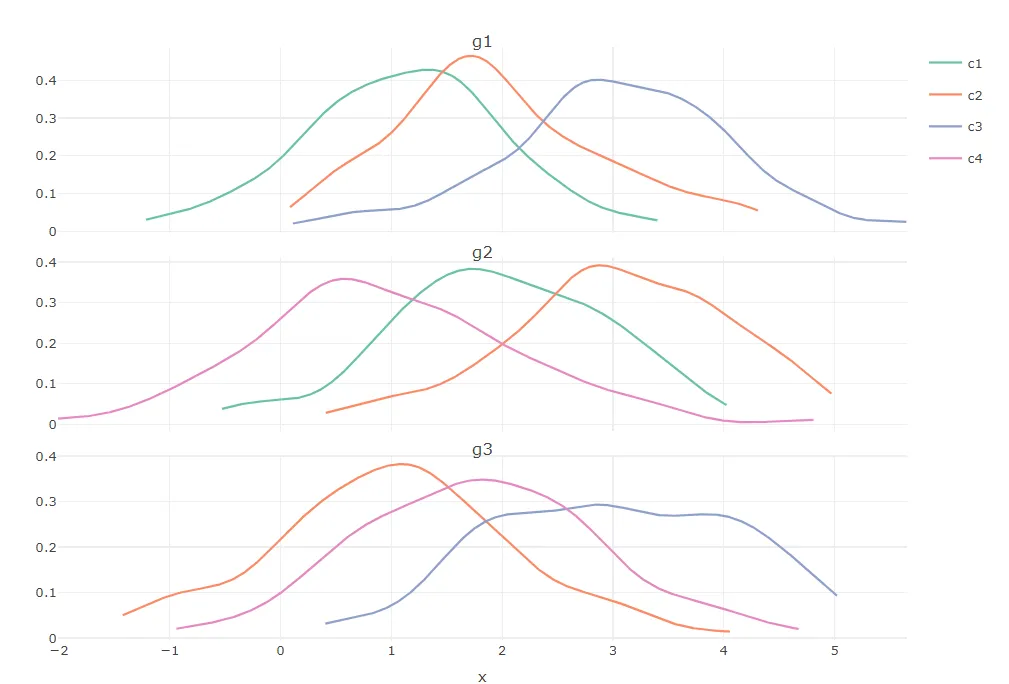
我有几个组,每个组都有几个类别,我对每个类别进行了连续值的测量:
set.seed(1)
df <- data.frame(value = c(rnorm(100,1,1), rnorm(100,2,1), rnorm(100,3,1),
rnorm(100,3,1), rnorm(100,1,1), rnorm(100,2,1),
rnorm(100,2,1), rnorm(100,3,1), rnorm(100,1,1)),
class = c(rep("c1",100), rep("c2",100), rep("c3",100),
rep("c2",100), rep("c4",100), rep("c1",100),
rep("c4",100), rep("c3",100), rep("c2",100)),
group = c(rep("g1",300), rep("g2",300), rep("g3",300)))
df$class <- factor(df$class, levels =c("c1","c2","c3","c4"))
df$group <- factor(df$group, levels =c("g1","g2","g3"))
数据中的每个组并不具有相同的类,或者说每个组都有所有类的子集。
我正在尝试生成针对每个组的 R plotly 密度曲线,按类别进行彩色编码,然后使用 plotly 的 subplot 函数将它们组合到一个单独的图中。
这就是我正在做的事情:
library(dplyr)
library(ggplot2)
library(plotly)
set.seed(1)
df <- data.frame(value = c(rnorm(100,1,1), rnorm(100,2,1), rnorm(100,3,1),
rnorm(100,3,1), rnorm(100,1,1), rnorm(100,2,1),
rnorm(100,2,1), rnorm(100,3,1), rnorm(100,1,1)),
class = c(rep("c1",100), rep("c2",100), rep("c3",100),
rep("c2",100), rep("c4",100), rep("c1",100),
rep("c4",100), rep("c3",100), rep("c2",100)),
group = c(rep("g1",300), rep("g2",300), rep("g3",300)))
df$class <- factor(df$class, levels =c("c1","c2","c3","c4"))
df$group <- factor(df$group, levels =c("g1","g2","g3"))
plot.list <- lapply(c("g1","g2","g3"), function(g){
density.df <- do.call(rbind,lapply(unique(dplyr::filter(df, group == g)$class),function(l)
ggplot_build(ggplot(dplyr::filter(df, group == g & class == l),aes(x=value))+geom_density(adjust=1,colour="#A9A9A9"))$data[[1]] %>%
dplyr::select(x,y) %>% dplyr::mutate(class = l)))
plot_ly(x = density.df$x, y = density.df$y, type = 'scatter', mode = 'lines',color = density.df$class) %>%
layout(title=g,xaxis = list(zeroline = F), yaxis = list(zeroline = F))
})
subplot(plot.list,nrows=length(plot.list),shareX=T)
这里给出:
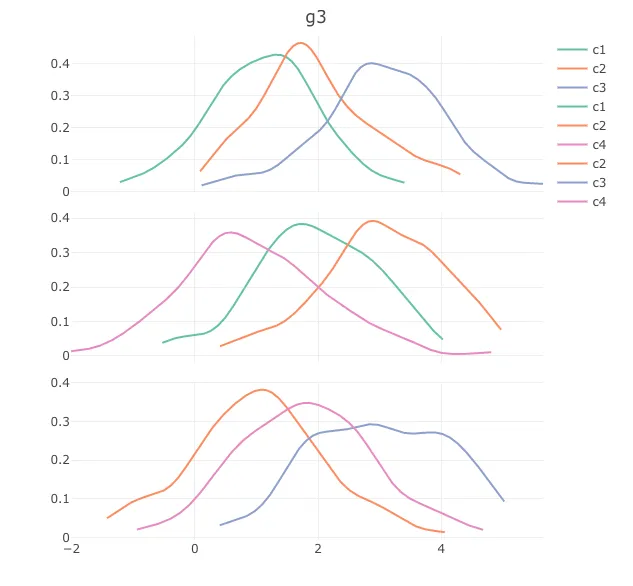
我想要解决的问题是:
- 只在图例中出现一次(现在对于每个组都会重复出现),合并所有类别
- 使标题在每个子图中出现,而不仅仅是像现在这样只在最后一个图中出现。(我知道可以将组名作为x轴标题,但我宁愿节省空间,因为实际上我有多于3个组)