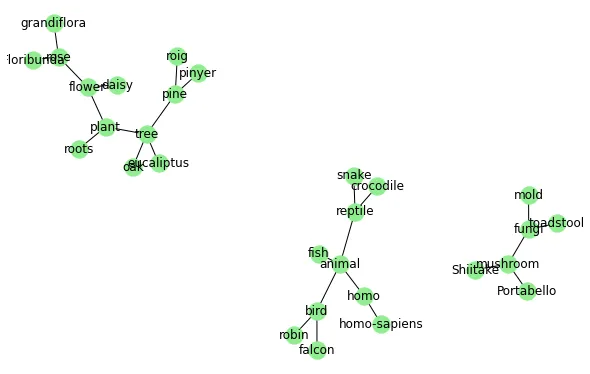
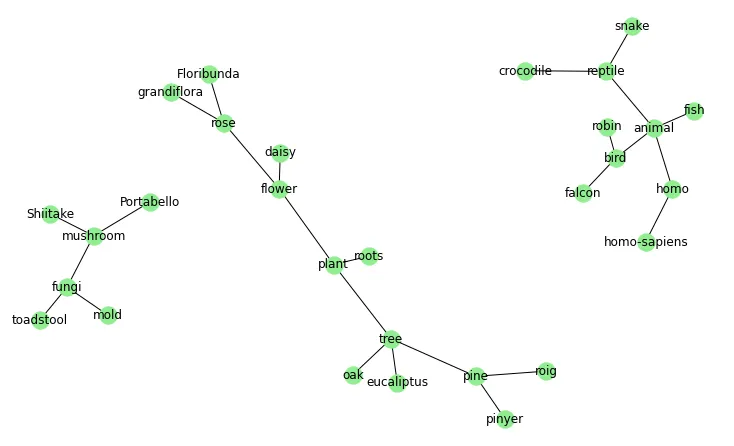
我使用networkx和plotly创建了以下网络:
我得到了这个图: 网络 我想添加标签,但是它变得太拥挤了(特别是在中间),因此我有两个问题:
edges = df.stack().reset_index()
edges.columns = ['var_1','var_2','correlation']
edges = edges.loc[ (edges['correlation'] < -0.6) | (edges['correlation'] > 0.6) & (edges['var_1'] != edges['var_2']) ].copy()
#create undirected graph with weights corresponding to the correlation magnitude
G0 = nx.from_pandas_edgelist(edges, 'var_1', 'var_2', edge_attr=['correlation'])
mst = G0
# assign colours to edges depending on positive or negative correlation
# assign edge thickness depending on magnitude of correlation
edge_colours = []
edge_width = []
for key, value in nx.get_edge_attributes(mst, 'correlation').items():
edge_colours.append(assign_colour(value))
edge_width.append(assign_thickness(value))
node_size = []
degrees = [val for key, val in dict(G0.degree).items()]
max_deg = max(degrees)
min_deg = min(degrees)
for value in degrees:
node_size.append(assign_node_size(value,min_deg,max_deg))
#draw the network:
nx.draw(mst, pos=nx.fruchterman_reingold_layout(mst),
node_size=15, edge_color=edge_colours, node_colour="black",
edge_width=0.2)
plt.show()
def get_coordinates(G=mst):
"""Returns the positions of nodes and edges in a format for Plotly to draw the network"""
# get list of node positions
pos = nx.fruchterman_reingold_layout(mst)
Xnodes = [pos[n][0] for n in mst.nodes()]
Ynodes = [pos[n][1] for n in mst.nodes()]
Xedges_red = []
Yedges_red = []
Xedges_green = []
Yedges_green = []
def insert_edge(Xedges, Yedges):
Xedges.extend([pos[e[0]][0], pos[e[1]][0], None])
Yedges.extend([pos[e[0]][1], pos[e[1]][1], None])
search_dict = nx.get_edge_attributes(mst, 'correlation')
for e in mst.edges():
correlation = search_dict[(e[0], e[1])]
if correlation <= 0 : # red_edges
insert_edge(Xedges_red, Yedges_red)
else:
insert_edge(Xedges_green, Yedges_green)
# x coordinates of the nodes defining the edge e
return Xnodes, Ynodes, Xedges_red, Yedges_red, Xedges_green, Yedges_green
node_label = list(mst.nodes())
node_label = [fix_string(x) for x in node_label]
# get coordinates for nodes and edges
Xnodes, Ynodes, Xedges_red, Yedges_red, Xedges_green, Yedges_green = get_coordinates()
external_data = [list(x) for x in coding_names_df.values]
external_data = {fix_string(x[0]): x[1] for x in external_data}
external_data2 = [list(y) for y in coding_names_df.values]
external_data2 = {fix_string(y[0]): y[2] for y in external_data2}
external_data3 = [list(z) for z in coding_names_df.values]
external_data3 = {fix_string(z[0]): z[3] for z in external_data3}
external_data4 = [list(s) for s in coding_names_df.values]
external_data4 = {fix_string(s[0]): s[4] for s in external_data4}
# =============================================================================
description = [f"<b>{index}) {node}</b>"
"<br><br>Realm: " +
"<br>" + external_data.get(node, 'No external data found') +
"<br><br>Type: " +
"<br>" + external_data2.get(node, 'No external data found')
for index, node in enumerate(node_label)]
# =============================================================================
# def nodes colours:
node_colour = [assign_node_colour(node, external_data3, coding_names_df) for node in node_label]
node_shape = [assign_node_shape(node, external_data4, coding_names_df) for node in node_label]
# edges
# negative:
tracer_red = go.Scatter(x=Xedges_red, y=Yedges_red,
mode='lines',
line= dict(color="#FA0000", width=1),
hoverinfo='none',
showlegend=False)
# positive:
tracer_green = go.Scatter(x=Xedges_green, y=Yedges_green,
mode='lines',
line= dict(color= "#29A401", width=1),
hoverinfo='none',
showlegend=False)
# nodes
tracer_marker = go.Scatter(x=Xnodes, y=Ynodes,
mode='markers+text',
textposition='top center',
marker=dict(size=node_size,
line=dict(width=0.8, color='black'),
color=node_colour,
symbol=node_shape),
hovertext=description,
hoverinfo='text',
textfont=dict(size=7),
showlegend=False)
axis_style = dict(title='',
titlefont=dict(size=20),
showgrid=False,
zeroline=False,
showline=False,
ticks='',
showticklabels=False)
layout = dict(title='',
width=1300,
height=900,
autosize=False,
showlegend=False,
xaxis=axis_style,
yaxis=axis_style,
hovermode='closest',
plot_bgcolor = '#fff')
fig = dict(data=[tracer_red, tracer_green, tracer_marker], layout=layout)
display(HTML("""<p>Node sizes are proportional to the size of annualised returns.<br>
Node colours signify positive or negative returns since beginning of the timeframe.</p> """))
plot(fig)
我得到了这个图: 网络 我想添加标签,但是它变得太拥挤了(特别是在中间),因此我有两个问题:
- 如何在中间空出位置? (但仍要保持fruchterman_reingold位置)
- 如何添加一些特定的标签?