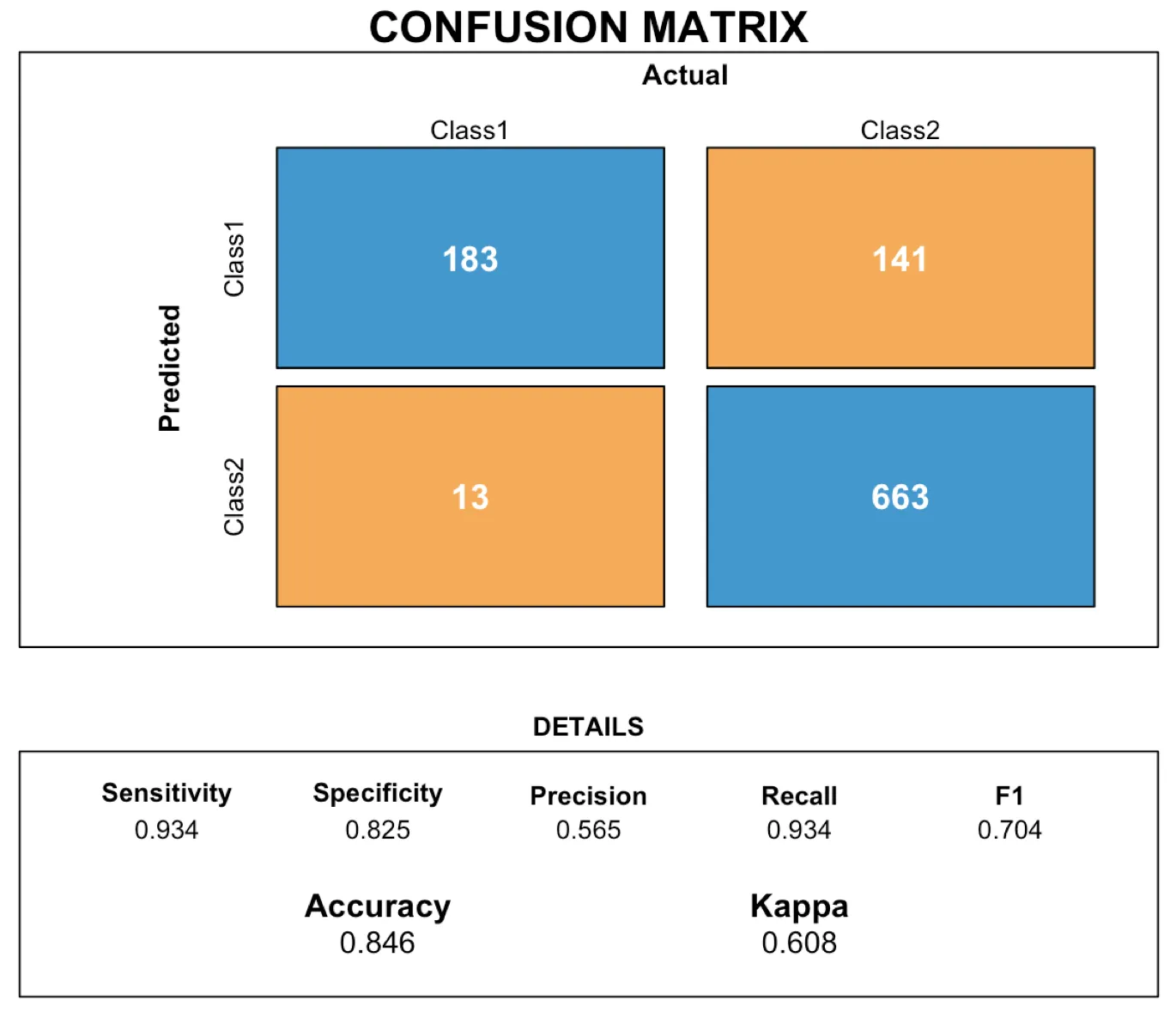
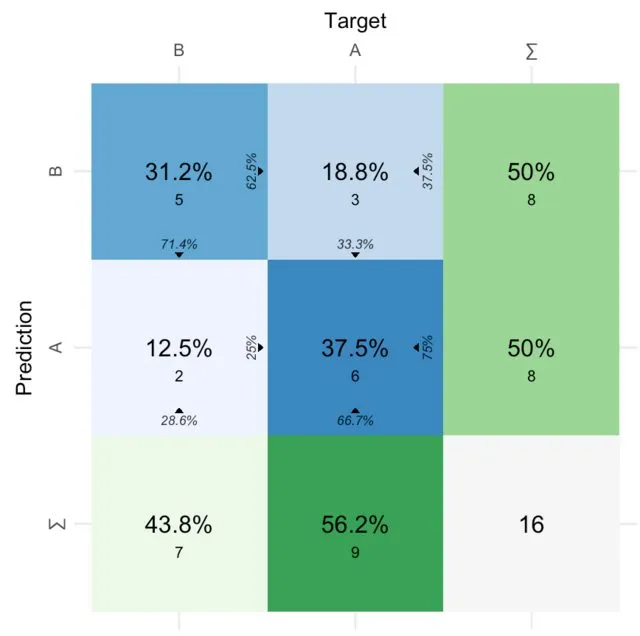
您可以在R中使用rect功能来布局混淆矩阵。在这里,我们将创建一个函数,允许用户传递由caret软件包创建的cm对象,以生成可视化结果。
让我们先创建一个评估数据集,正如在caret演示中所做的那样。
set.seed(144)
true_class <- factor(sample(paste0("Class", 1:2), size = 1000, prob = c(.2, .8), replace = TRUE))
true_class <- sort(true_class)
class1_probs <- rbeta(sum(true_class == "Class1"), 4, 1)
class2_probs <- rbeta(sum(true_class == "Class2"), 1, 2.5)
test_set <- data.frame(obs = true_class,Class1 = c(class1_probs, class2_probs))
test_set$Class2 <- 1 - test_set$Class1
test_set$pred <- factor(ifelse(test_set$Class1 >= .5, "Class1", "Class2"))
现在让我们使用caret来计算混淆矩阵:
# calculate the confusion matrix
cm <- confusionMatrix(data = test_set$pred, reference = test_set$obs)
现在我们创建一个函数,根据需要布置矩形,以更具视觉吸引力的方式展示混淆矩阵:
draw_confusion_matrix <- function(cm) {
layout(matrix(c(1,1,2)))
par(mar=c(2,2,2,2))
plot(c(100, 345), c(300, 450), type = "n", xlab="", ylab="", xaxt='n', yaxt='n')
title('CONFUSION MATRIX', cex.main=2)
rect(150, 430, 240, 370, col='#3F97D0')
text(195, 435, 'Class1', cex=1.2)
rect(250, 430, 340, 370, col='#F7AD50')
text(295, 435, 'Class2', cex=1.2)
text(125, 370, 'Predicted', cex=1.3, srt=90, font=2)
text(245, 450, 'Actual', cex=1.3, font=2)
rect(150, 305, 240, 365, col='#F7AD50')
rect(250, 305, 340, 365, col='#3F97D0')
text(140, 400, 'Class1', cex=1.2, srt=90)
text(140, 335, 'Class2', cex=1.2, srt=90)
res <- as.numeric(cm$table)
text(195, 400, res[1], cex=1.6, font=2, col='white')
text(195, 335, res[2], cex=1.6, font=2, col='white')
text(295, 400, res[3], cex=1.6, font=2, col='white')
text(295, 335, res[4], cex=1.6, font=2, col='white')
plot(c(100, 0), c(100, 0), type = "n", xlab="", ylab="", main = "DETAILS", xaxt='n', yaxt='n')
text(10, 85, names(cm$byClass[1]), cex=1.2, font=2)
text(10, 70, round(as.numeric(cm$byClass[1]), 3), cex=1.2)
text(30, 85, names(cm$byClass[2]), cex=1.2, font=2)
text(30, 70, round(as.numeric(cm$byClass[2]), 3), cex=1.2)
text(50, 85, names(cm$byClass[5]), cex=1.2, font=2)
text(50, 70, round(as.numeric(cm$byClass[5]), 3), cex=1.2)
text(70, 85, names(cm$byClass[6]), cex=1.2, font=2)
text(70, 70, round(as.numeric(cm$byClass[6]), 3), cex=1.2)
text(90, 85, names(cm$byClass[7]), cex=1.2, font=2)
text(90, 70, round(as.numeric(cm$byClass[7]), 3), cex=1.2)
text(30, 35, names(cm$overall[1]), cex=1.5, font=2)
text(30, 20, round(as.numeric(cm$overall[1]), 3), cex=1.4)
text(70, 35, names(cm$overall[2]), cex=1.5, font=2)
text(70, 20, round(as.numeric(cm$overall[2]), 3), cex=1.4)
}
最后,将我们使用caret计算混淆矩阵时计算出的cm对象传入:
draw_confusion_matrix(cm)
以下是结果: