我正在使用半方差函数研究我的数据中的空间自相关性。我的数据:
Response <- c(21L, 36L, 30L, 29L, 30L, 45L, 100L, 0L, 0L, 0L, 0L, 0L, 59L,
18L, 24L, 23L, 26L, 29L, 23L, 21L, 14L, 30L, 43L, 14L, 8L, 0L,
0L, 0L, 0L, 0L, 23L, 38L, 20L, 28L, 45L, 21L, 46L, 23L, 6L, 4L,
0L, 0L, 0L, 0L, 0L, 17L, 10L, 41L, 24L, 31L, 16L, 23L, 31L, 6L,
2L, 0L, 0L, 0L, 0L, 0L, 8L, 20L, 18L, 18L, 40L, 9L, 1L, 25L,
4L, 34L, 0L, 0L, 0L, 0L, 0L, 39L, 8L, 7L, 22L, 16L, 18L, 23L,
11L, 25L, 28L, 0L, 0L, 0L, 0L, 0L, 3L, 22L, 11L, 9L, 123L, 50L,
12L, 1L, 46L, 1L, 4L, 1L, 2L, 0L, 37L)
Covar1 <- structure(c(1L, 3L, 1L, 1L, 3L, 3L, 1L, 2L, 2L, 2L, 2L, 2L, 1L,
3L, 3L, 1L, 3L, 1L, 1L, 3L, 3L, 1L, 1L, 3L, 3L, 2L, 2L, 2L, 2L,
2L, 1L, 3L, 1L, 1L, 3L, 3L, 1L, 1L, 3L, 3L, 2L, 2L, 2L, 2L, 2L,
1L, 3L, 1L, 1L, 3L, 3L, 1L, 1L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 1L,
3L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 1L, 2L, 2L, 2L, 2L, 2L, 1L, 3L,
3L, 3L, 3L, 1L, 1L, 3L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 1L,
3L, 1L, 1L, 3L, 3L, 1L, 2L, 2L, 2L, 2L, 2L, 1L), .Label = c("A",
"B", "C"), class = "factor")
Covar2 <- structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 5L,
5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 7L, 7L, 7L,
7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L), .Label = c("1",
"2", "3", "4", "5", "6", "7"), class = "factor")
df <- data.frame(Response, Covar1, Covar2)
我运行了一个简单的模型,使用
gstat从残差和空间坐标中制作经验和拟合半变异函数,并将它们绘制出来:mod1 <- glm(Response ~ Covar1 * Covar2, data = df)
geo <- as.data.frame(resid(mod1))
geo$x <- c(34.59481, 34.60548, 34.59825, 34.59039, 34.56546, 34.56749,
34.5964, 34.40986, 34.40083, 34.39536, 34.41291, 34.40512, 34.36381,
34.35335102, 34.32548, 34.59481, 34.60548, 34.59825, 34.59039,
34.56749, 34.56546, 34.5964, 34.36381, 34.35335102, 34.32548,
34.41291, 34.40986, 34.40512, 34.40083, 34.39536, 34.59481, 34.60548,
34.59825, 34.59039, 34.56749, 34.56546, 34.5964, 34.36381, 34.35335102,
34.32548, 34.41291, 34.40986, 34.40512, 34.40083, 34.39536, 34.59481,
34.60548, 34.59825, 34.59039, 34.56749, 34.56546, 34.5964, 34.36381,
34.35335102, 34.32548, 34.41291, 34.40986, 34.40512, 34.40083,
34.39536, 34.59481, 34.60548, 34.59825, 34.59039, 34.36381, 34.35335102,
34.32548, 34.56749, 34.56546, 34.5964, 34.41291, 34.40986, 34.40512,
34.40083, 34.39536, 34.36381, 34.35335102, 34.32548, 34.56546,
34.56749, 34.5964, 34.59481, 34.60548, 34.59825, 34.59039, 34.41291,
34.40986, 34.40512, 34.40083, 34.39536, 34.32548, 34.35335102,
34.59481, 34.60548, 34.59039, 34.59825, 34.56546, 34.56749, 34.5964,
34.41291, 34.40986, 34.40512, 34.40083, 34.39536, 34.36381)
geo$y <- c(-2.18762, -2.18308, -2.16174, -2.16018, -2.14787, -2.15296,
-2.12863, -2.14325, -2.14552, -2.1454, -2.13926, -2.14652, -2.12463,
-2.121925978, -2.10213, -2.18762, -2.18308, -2.16174, -2.16018,
-2.15296, -2.14787, -2.12863, -2.12463, -2.121925978, -2.10213,
-2.13926, -2.14325, -2.14652, -2.14552, -2.1454, -2.18762, -2.18308,
-2.16174, -2.16018, -2.15296, -2.14787, -2.12863, -2.12463, -2.121925978,
-2.10213, -2.13926, -2.14325, -2.14652, -2.14552, -2.1454, -2.18762,
-2.18308, -2.16174, -2.16018, -2.15296, -2.14787, -2.12863, -2.12463,
-2.121925978, -2.10213, -2.13926, -2.14325, -2.14652, -2.14552,
-2.1454, -2.18762, -2.18308, -2.16174, -2.16018, -2.12463, -2.121925978,
-2.10213, -2.15296, -2.14787, -2.12863, -2.13926, -2.14325, -2.14652,
-2.14552, -2.1454, -2.12463, -2.121925978, -2.10213, -2.14787,
-2.15296, -2.12863, -2.18762, -2.18308, -2.16174, -2.16018, -2.13926,
-2.14325, -2.14652, -2.14552, -2.1454, -2.10213, -2.121925978,
-2.18762, -2.18308, -2.16018, -2.16174, -2.14787, -2.15296, -2.12863,
-2.13926, -2.14325, -2.14652, -2.14552, -2.1454, -2.12463)
library(sp)
names(geo) <- c("resids", "x", "y")
coordinates(geo) <- ~ x + y
proj4string(geo) <- CRS("+proj=longlat +datum=WGS84")
library(gstat)
var1 <- variogram(resids ~ x + y, data = geo)
v.fit1 = fit.variogram(var1, vgm(50, "Exp", 2, 50))
plot(var1, v.fit1)
ggplot(var1, aes(x=dist,y=gamma)) +
geom_point()
但是我在绘制拟合模型(即线条)时遇到了困难。非常感谢任何帮助。
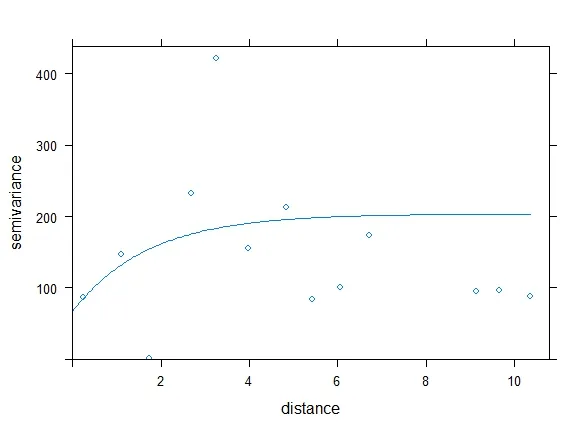
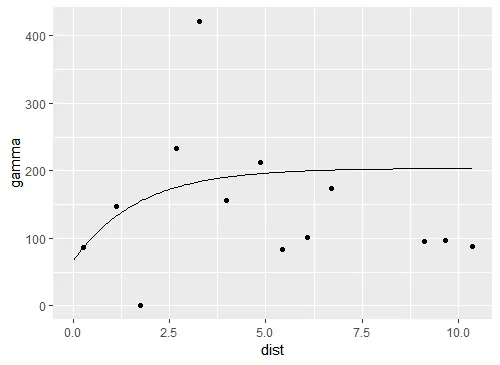

geom_smooth(method="glm")或"lm"、"gam"、"loess"、"rlm"。 - Roman