我希望使用
ggplot2画出以下drc::plot.drc的图形。
df1 <-
structure(list(TempV = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 5L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L, 7L,
7L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 9L, 13L, 13L, 13L, 13L,
13L, 13L, 13L, 13L, 13L, 13L, 11L, 11L, 11L, 11L, 11L, 11L, 11L,
11L, 11L, 11L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 8L, 10L, 10L, 10L,
10L, 10L, 10L, 10L, 10L, 10L, 10L, 14L, 14L, 14L, 14L, 14L, 14L,
14L, 14L, 14L, 14L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L, 12L,
12L), .Label = c("22.46FH-142", "27.59FH-142", "26.41FH-142",
"29.71FH-142", "31.66FH-142", "34.11FH-142", "33.22FH-142", "22.46FH-942",
"27.59FH-942", "26.41FH-942", "29.71FH-942", "31.66FH-942", "34.11FH-942",
"33.22FH-942"), class = "factor"), Start = c(0L, 24L, 48L, 72L,
96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L,
144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L, 144L, 168L,
192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L, 144L, 168L, 192L, 216L,
0L, 24L, 48L, 72L, 96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L,
48L, 72L, 96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L,
96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L,
144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L, 144L, 168L,
192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L, 144L, 168L, 192L, 216L,
0L, 24L, 48L, 72L, 96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L,
48L, 72L, 96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L,
96L, 120L, 144L, 168L, 192L, 216L, 0L, 24L, 48L, 72L, 96L, 120L,
144L, 168L, 192L, 216L), End = c(24, 48, 72, 96, 120, 144, 168,
192, 216, Inf, 24, 48, 72, 96, 120, 144, 168, 192, 216, Inf,
24, 48, 72, 96, 120, 144, 168, 192, 216, Inf, 24, 48, 72, 96,
120, 144, 168, 192, 216, Inf, 24, 48, 72, 96, 120, 144, 168,
192, 216, Inf, 24, 48, 72, 96, 120, 144, 168, 192, 216, Inf,
24, 48, 72, 96, 120, 144, 168, 192, 216, Inf, 24, 48, 72, 96,
120, 144, 168, 192, 216, Inf, 24, 48, 72, 96, 120, 144, 168,
192, 216, Inf, 24, 48, 72, 96, 120, 144, 168, 192, 216, Inf,
24, 48, 72, 96, 120, 144, 168, 192, 216, Inf, 24, 48, 72, 96,
120, 144, 168, 192, 216, Inf, 24, 48, 72, 96, 120, 144, 168,
192, 216, Inf, 24, 48, 72, 96, 120, 144, 168, 192, 216, Inf),
Germinated = c(0L, 0L, 0L, 0L, 3L, 67L, 46L, 12L, 101L, 221L,
0L, 0L, 0L, 0L, 57L, 50L, 44L, 31L, 32L, 236L, 0L, 0L, 0L,
31L, 68L, 50L, 31L, 34L, 29L, 207L, 0L, 0L, 8L, 30L, 31L,
55L, 27L, 22L, 4L, 273L, 0L, 0L, 46L, 64L, 16L, 8L, 15L,
15L, 20L, 266L, 0L, 0L, 0L, 0L, 4L, 13L, 63L, 51L, 147L,
172L, 0L, 0L, 4L, 26L, 92L, 31L, 91L, 14L, 7L, 185L, 0L,
0L, 0L, 0L, 0L, 32L, 59L, 36L, 50L, 273L, 0L, 0L, 0L, 4L,
13L, 32L, 42L, 52L, 42L, 265L, 0L, 0L, 0L, 6L, 22L, 40L,
57L, 44L, 73L, 208L, 0L, 1L, 2L, 24L, 55L, 41L, 68L, 24L,
33L, 202L, 0L, 0L, 18L, 31L, 26L, 30L, 61L, 25L, 58L, 201L,
0L, 0L, 36L, 54L, 33L, 55L, 12L, 27L, 55L, 178L, 0L, 0L,
6L, 28L, 26L, 31L, 53L, 48L, 33L, 225L)), .Names = c("TempV",
"Start", "End", "Germinated"), row.names = c(NA, -140L), class = "data.frame")
library(data.table)
dt1 <- data.table(df1)
library(drc)
dt1fm1 <-
drm(
formula = Germinated ~ Start + End
, curveid = TempV
# , pmodels =
# , weights =
, data = dt1
# , subset =
, fct = LL.2()
, type = "event"
, bcVal = NULL
, bcAdd = 0
# , start =
, na.action = na.fail
, robust = "mean"
, logDose = NULL
, control = drmc(
constr = FALSE
, errorm = TRUE
, maxIt = 1500
, method = "BFGS"
, noMessage = FALSE
, relTol = 1e-07
, rmNA = FALSE
, useD = FALSE
, trace = FALSE
, otrace = FALSE
, warnVal = -1
, dscaleThres = 1e-15
, rscaleThres = 1e-15
)
, lowerl = NULL
, upperl = NULL
, separate = FALSE
, pshifts = NULL
)
## ----dt1fm1Plot1----
plot(
x = dt1fm1
, xlab = "Time (Hours)"
, ylab = "Proportion Germinated (\\%)"
# , ylab = "Proportion Germinated (%)"
, add = FALSE
, level = NULL
, type = "average" # c("average", "all", "bars", "none", "obs", "confidence")
, broken = FALSE
# , bp
, bcontrol = NULL
, conName = NULL
, axes = TRUE
, gridsize = 100
, log = ""
# , xtsty
, xttrim = TRUE
, xt = NULL
, xtField = NULL
, xField = "Time (Hours)"
, xlim = c(0, 200)
, yt = NULL
, ytField = NULL
, yField = "Proportion Germinated"
, ylim = c(0, 1.05)
, lwd = 1
, cex = 1.2
, cex.axis = 1
, col = TRUE
# , lty
# , pch
, legend = TRUE
# , legendText
, legendPos = c(40, 1.1)
, cex.legend = 0.6
, normal = FALSE
, normRef = 1
, confidence.level = 0.95
)
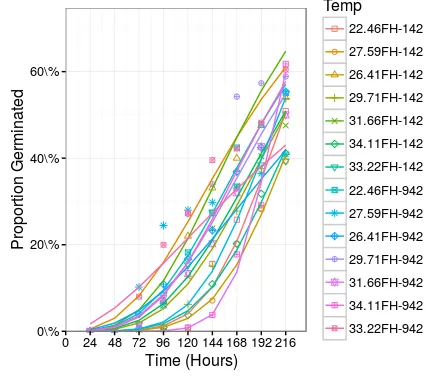
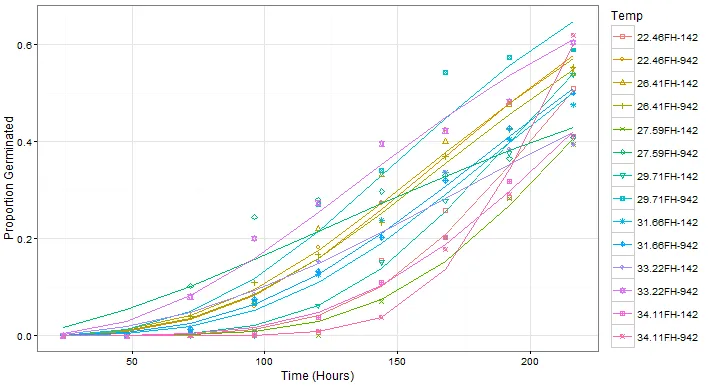
## ----dt1fm1Plot2----
dt1fm1Means1 <- dt1[, .(Germinated=mean(Germinated)/450), by=.(TempV, Start, End)]
dt1fm1Means2 <- dt1fm1Means1[, .(Start=Start, End=End, Cum_Germinated=cumsum(Germinated)), by=.(TempV)]
dt1fm1Means <- data.table(dt1fm1Means2[End!=Inf], Pred=predict(object=dt1fm1))
dt1fm1Plot2 <-
ggplot(data= dt1fm1Means, mapping=aes(x=End, y=Cum_Germinated, group=TempV, color=TempV, shape=TempV)) +
geom_point() +
geom_line(aes(y = Pred)) +
scale_shape_manual(values=seq(0, 13)) +
labs(x = "Time (Hours)", y = "Proportion Germinated", shape="Temp", color="Temp") +
theme_bw() +
scale_x_continuous(expand = c(0, 0), breaks = c(0, unique(dt1fm1Means$End))) +
scale_y_continuous(expand = c(0, 0), labels = function(x) paste0(100*x,"\\%")) +
# scale_y_continuous(expand = c(0, 0), labels = percent) +
expand_limits(x = c(0, max(dt1fm1Means$End)+20), y = c(0, max(dt1fm1Means$Pred)+0.1)) +
theme(axis.title.x = element_text(size = 12, hjust = 0.54, vjust = 0),
axis.title.y = element_text(size = 12, angle = 90, vjust = 0.25))
print(dt1fm1Plot2)
问题
ggplot2 输出中存在一些差异。这些差异是由于 predict 函数给出的输出与数据中给定的水平不同。
编辑后
实际上,drm 函数改变了 TempV 的水平顺序,这在 summary(dt1fm1) 输出和 drc::plot.drc 输出的图表中很清楚。
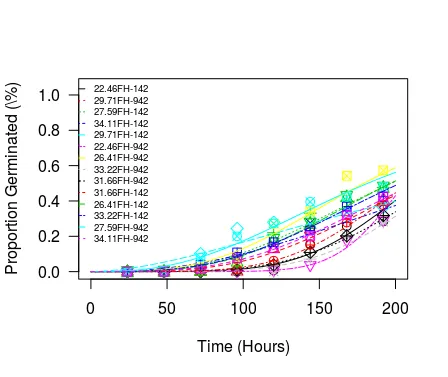
ggplot2为一个曲线绘制drc模型(请参见此处)。然而,当使用curveid = TempV时,我遇到了困难。有什么想法吗? - MYaseen208