我已经对一些值进行了聚类,然后将它们分组。接着,我使用ggplot2绘制了一些密度图并将聚类结果叠加在上面。下面是一个示例图像:
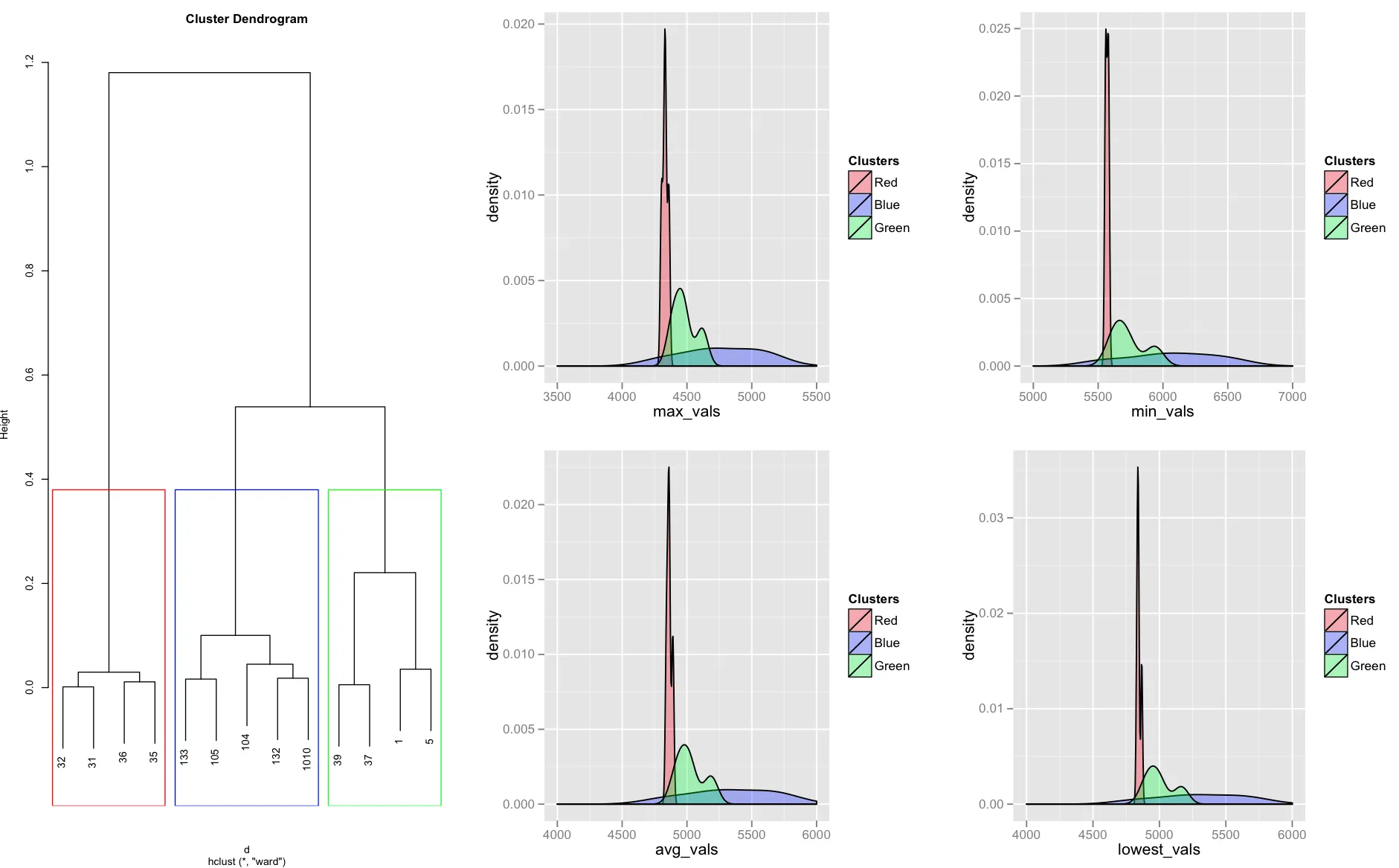
对于聚类中的每个组,我都会绘制一个密度图并将它们叠加在一起。密度图中的颜色对应于聚类中的分组。
我的问题是,我手动根据分组拆分了数据,并将其放入各自的文本表格中(请参见下面的代码)。这非常低效,对于大型数据集来说可能非常繁琐。如何在不将聚类分离为各自的文本表格的情况下,在ggplot2中动态绘制密度图?
在拆分之前,原始输入表格如下所示:
scores <- read.table(textConnection("
file max min avg lowest
132 5112.0 6520.0 5728.0 5699.0
133 4720.0 6064.0 5299.0 5277.0
5 4617.0 5936.0 5185.0 5165.0
1 4384.0 5613.0 4917.0 4895.0
1010 5008.0 6291.0 5591.0 5545.0
104 4329.0 5554.0 4858.0 4838.0
105 4636.0 5905.0 5193.0 5165.0
35 4304.0 5578.0 4842.0 4831.0
36 4360.0 5580.0 4891.0 4867.0
37 4444.0 5663.0 4979.0 4952.0
31 4328.0 5559.0 4858.0 4839.0
39 4486.0 5736.0 5031.0 5006.0
32 4334.0 5558.0 4864.0 4843.0
"), header=TRUE)
我使用的代码生成绘图: 请注意,将基本图形与网格组合仍然无法正常工作。
library(ggplot2)
library(grid)
layout(matrix(c(1,2,3,1,4,5), 2, 3, byrow = TRUE))
# define function to create multi-plot setup (nrow, ncol)
vp.setup <- function(x,y){
grid.newpage()
pushViewport(viewport(layout = grid.layout(x,y)))
}
# define function to easily access layout (row, col)
vp.layout <- function(x,y){
viewport(layout.pos.row=x, layout.pos.col=y)
}
vp.setup(2,3)
file_vals <- read.table(textConnection("
file avg_vals
133 1.5923
132 1.6351
1010 1.6532
104 1.6824
105 1.6087
39 1.8694
32 1.9934
31 1.9919
37 1.8638
36 1.9691
35 1.9802
1 1.7283
5 1.7637
"), header=TRUE)
red <- read.table(textConnection("
file max min avg lowest
31 4328.0 5559.0 4858.0 4839.0
32 4334.0 5558.0 4864.0 4843.0
36 4360.0 5580.0 4891.0 4867.0
35 4304.0 5578.0 4842.0 4831.0
"), header=TRUE)
blue <- read.table(textConnection("
file max min avg lowest
133 4720.0 6064.0 5299.0 5277.0
105 4636.0 5905.0 5193.0 5165.0
104 4329.0 5554.0 4858.0 4838.0
132 5112.0 6520.0 5728.0 5699.0
1010 5008.0 6291.0 5591.0 5545.0
"), header=TRUE)
green <- read.table(textConnection("
file max min avg lowest
39 4486.0 5736.0 5031.0 5006.0
37 4444.0 5663.0 4979.0 4952.0
5 4617.0 5936.0 5185.0 5165.0
1 4384.0 5613.0 4917.0 4895.0
"), header=TRUE)
# Perform Cluster
d <- dist(file_vals$avg_vals, method = "euclidean")
fit <- hclust(d, method="ward")
plot(fit, labels=file_vals$file)
groups <- cutree(fit, k=3)
cols = c('red', 'blue', 'green', 'purple', 'orange', 'magenta', 'brown', 'chartreuse4','darkgray','cyan1')
rect.hclust(fit, k=3, border=cols)
# Desnity plots
dat = rbind(data.frame(Cluster='Red', max_vals = red$max), data.frame(Cluster='Blue', max_vals = blue$max), data.frame(Cluster='Green', max_vals = green$max))
max = (ggplot(dat,aes(x=max_vals)))
max = max + geom_density(aes(fill=factor(Cluster)), alpha=.3) + xlim(c(3500, 5500)) + scale_fill_manual(values=c("red",'blue',"green"))
max = max + labs(fill = 'Clusters')
print(max, vp=vp.layout(1,2))
dat = rbind(data.frame(Cluster='Red', min_vals = red$min), data.frame(Cluster='Blue', min_vals = blue$min), data.frame(Cluster='Green', min_vals = green$min))
min = (ggplot(dat,aes(x=min_vals)))
min = min + geom_density(aes(fill=factor(Cluster)), alpha=.3) + xlim(c(5000, 7000)) + scale_fill_manual(values=c("red",'blue',"green"))
min = min + labs(fill = 'Clusters')
print(min, vp=vp.layout(1,3))
dat = rbind(data.frame(Cluster='Red', avg_vals = red$avg), data.frame(Cluster='Blue', avg_vals = blue$avg), data.frame(Cluster='Green', avg_vals = green$avg))
avg = (ggplot(dat,aes(x=avg_vals)))
avg = avg + geom_density(aes(fill=factor(Cluster)), alpha=.3) + xlim(c(4000, 6000)) + scale_fill_manual(values=c("red",'blue',"green"))
avg = avg + labs(fill = 'Clusters')
print(avg, vp=vp.layout(2,2))
dat = rbind(data.frame(Cluster='Red', lowest_vals = red$lowest), data.frame(Cluster='Blue', lowest_vals = blue$lowest), data.frame(Cluster='Green', lowest_vals = green$lowest))
lowest = (ggplot(dat,aes(x=lowest_vals)))
lowest = lowest + geom_density(aes(fill=factor(Cluster)), alpha=.3) + xlim(c(4000, 6000)) + scale_fill_manual(values=c("red",'blue',"green"))
lowest = lowest + labs(fill = 'Clusters')
print(lowest, vp=vp.layout(2,3))
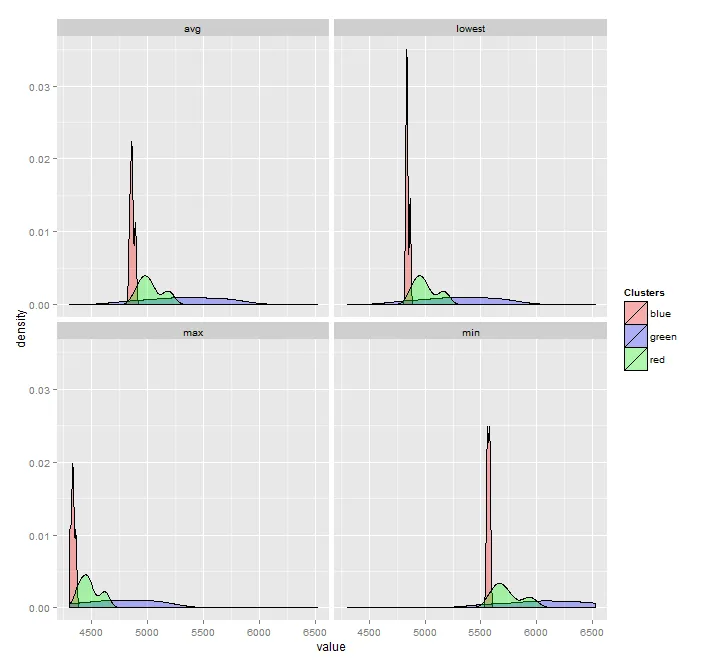
facet_wrap的另一个参数,基于每个面板数据的范围来允许x轴范围:facet_wrap(~ time, scales = "free_x")。请注意,使用ggplot2无法为面板指定不同的x轴。如果所有面板都需要相同的x轴,则可以将以下内容添加到您的图表中:+ coord_cartesian(xlim = c(3500, 7000))。 - Sven Hohenstein