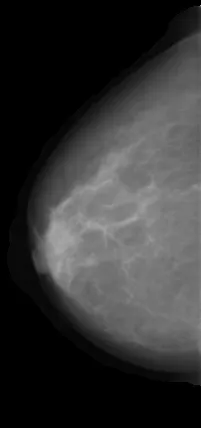
我有一个乳腺X光图像数据集(mini DDSM)。这些图片展示字母图案,指示左或右乳房,以及其他对我的机器学习模型无用的信息,因此我想在训练模型之前筛选这个数据集。
在本文中,他们使用Otsu二值化和开运算来清洁乳腺X光图像(第5/10页),链接如下:Preprocessing of Digital Mammogram Image Based on Otsu’s Threshold。
以下是他们的结果: 到目前为止,我已经编写了以下代码:
结果:
在本文中,他们使用Otsu二值化和开运算来清洁乳腺X光图像(第5/10页),链接如下:Preprocessing of Digital Mammogram Image Based on Otsu’s Threshold。
以下是他们的结果: 到目前为止,我已经编写了以下代码:
im = io.imread('/content/drive/MyDrive/TFM/DDSMPNG/ALL2/0.jpg')
# thresholding
thresh = im > filters.threshold_otsu(im)
# opening with a disk structure
disk = morphology.disk(5)
opened = morphology.binary_opening(thresh,disk)
# plotting
plt.figure(figsize=(10, 10))
plt.subplot(131)
plt.imshow(im,cmap='gray')
plt.subplot(132)
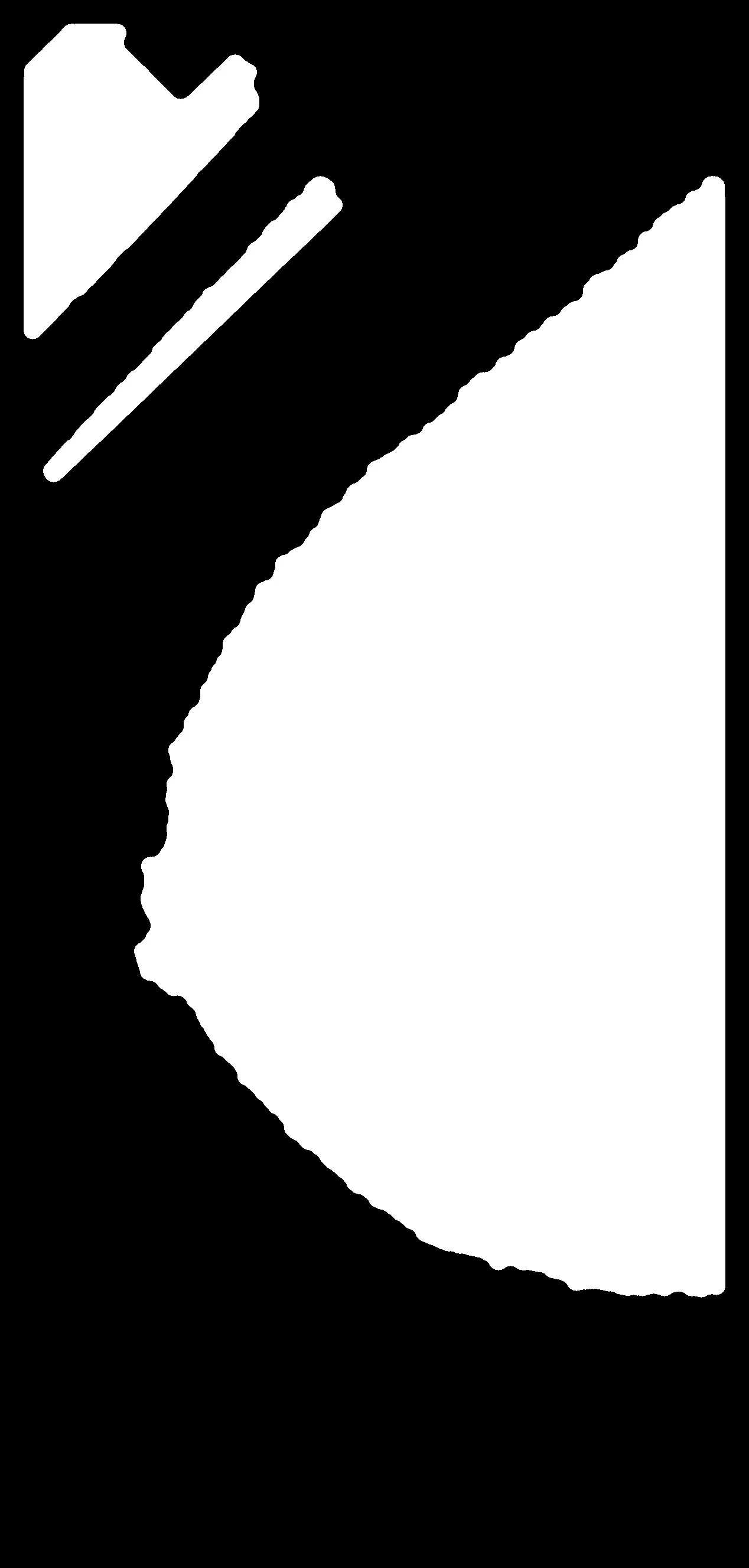
plt.imshow(opened,cmap='gray')
plt.imsave('/content/drive/MyDrive/TFM/DDSMPNG/Blackened/0.jpg',opened)
以下是图表:
我还尝试了更高的磁盘形状来做开运算,它似乎能够去除一些小字母形状的白色区域,但也会对乳腺X线照片进行一定程度的裁剪:
disk = morphology.disk(45)
opened = morphology.binary_opening(thresh,disk)
结果:
我猜我需要用二值化创建某种掩膜,并将其应用于原始图像,但我对图像处理库并不熟悉,也不确定如何实现这些结果。
编辑1:我尝试了 @fmw42 的建议,但遇到了一些问题(我在 Google Colab 上工作,不知道是否有关系...):
首先,在你的代码中使用的示例图像上,它似乎无法正常工作,不知道为什么,我只是修改了图像路径以及一些子绘图以查看结果:
# read image
img = cv2.imread('/content/drive/MyDrive/TFM/DDSMPNG/ALL2/0.jpg')
hh, ww = img.shape[:2]
# convert to grayscale
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# apply otsu thresholding
thresh = cv2.threshold(gray, 0, 255, cv2.THRESH_OTSU )[1]
# apply morphology close to remove small regions
kernel = cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (5,5))
morph = cv2.morphologyEx(thresh, cv2.MORPH_CLOSE, kernel)
# apply morphology open to separate breast from other regions
kernel = cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (5,5))
morph = cv2.morphologyEx(morph, cv2.MORPH_OPEN, kernel)
# get largest contour
contours = cv2.findContours(morph, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE)
contours = contours[0] if len(contours) == 2 else contours[1]
big_contour = max(contours, key=cv2.contourArea)
# draw largest contour as white filled on black background as mask
mask = np.zeros((hh,ww), dtype=np.uint8)
cv2.drawContours(mask, [big_contour], 0, 255, cv2.FILLED)
# dilate mask
kernel = cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (55,55))
mask = cv2.morphologyEx(mask, cv2.MORPH_DILATE, kernel)
# apply mask to image
result = cv2.bitwise_and(img, img, mask=mask)
# save results
cv2.imwrite('/content/drive/MyDrive/TFM/DDSMPNG/Blackened/0.jpg', result)
# show resultls
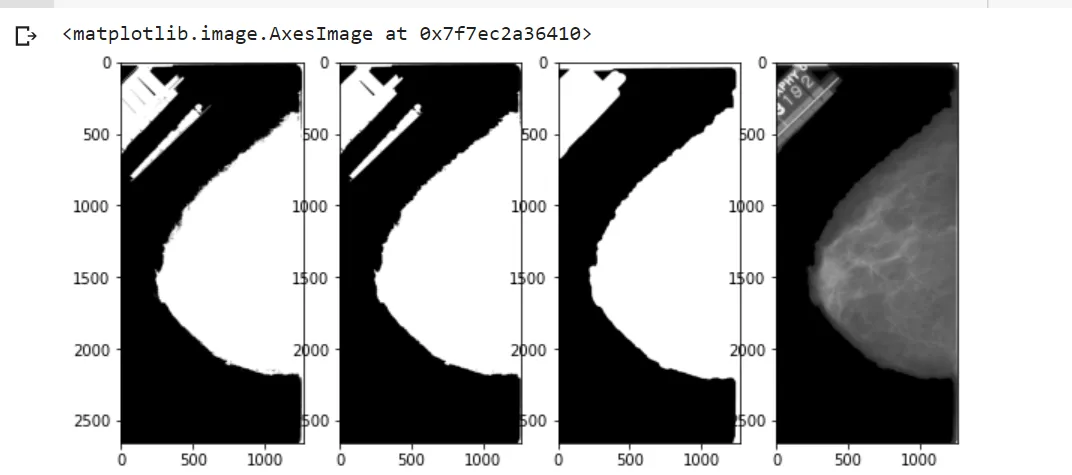
plt.figure(figsize=(10, 10))
plt.subplot(141)
plt.imshow(thresh,cmap='gray')
plt.subplot(142)
plt.imshow(morph,cmap='gray')
plt.subplot(143)
plt.imshow(mask,cmap='gray')
plt.subplot(144)
plt.imshow(result,cmap='gray')
结果:
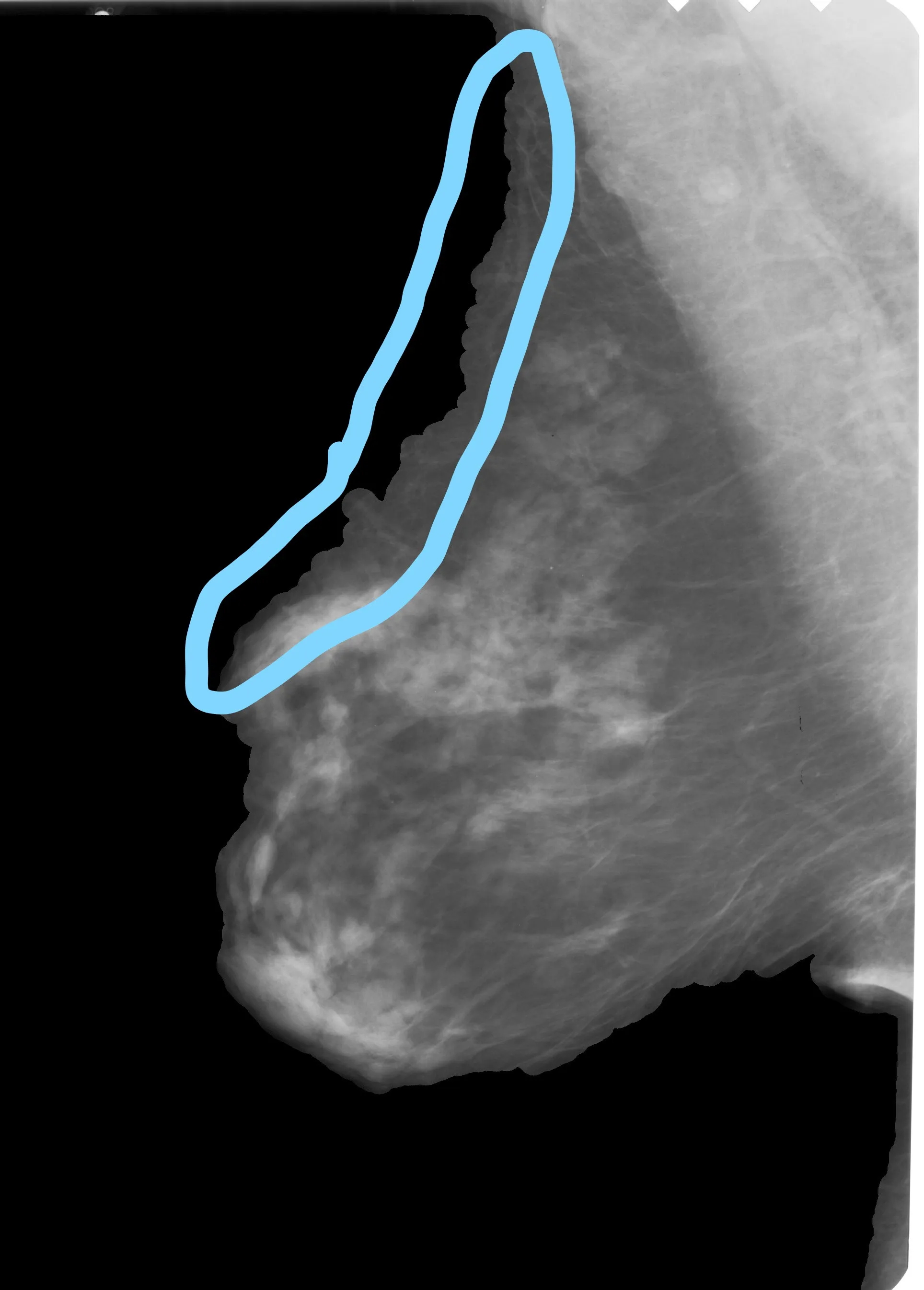
其次,对于其余的图像,它似乎对大多数图像都有效,但是会剪切一些乳房表面:
在您的结果图像中,它似乎更加平滑,我该如何实现它?
先行致谢!
编辑2:@fmw42的解决方案很好用,如果有人遇到相同的问题,您只需要调整形态学滤波器的核大小,直到图像的表现方式与他在答案中的结果相似。
非常感谢!