我认为您的错误来自于如何调用predict。我无法修复您的确切代码,但这里有一种简单的方法可以从您的模型中获取预测结果。使用purrr和nest的更复杂方法在此处进行了概述:
http://ijlyttle.github.io/isugg_purrr/presentation.html#(1)
更新-使用purrr和nest的方法
只需添加这个以展示在tidyverse中可以很容易地完成,使用predict即可。有关更多详细信息,请参见上面的链接。
library(tidyverse)
set.seed(1234)
myiris <- iris[sample(nrow(iris), replace = F),]
iris_nested <-
myiris[1:50,] %>%
nest(-Species) %>%
rename(myorigdata = data)
new_iris_nested <-
myiris[51:150,] %>%
nest(-Species) %>%
rename(mynewdata = data)
my_rlm <- function(df) {
MASS::rlm(Sepal.Length ~ Petal.Length + Petal.Width, data = df)
}
predictions_tall <-
iris_nested %>%
mutate(my_model = map(myorigdata, my_rlm)) %>%
full_join(new_iris_nested, by = "Species") %>%
mutate(my_new_pred = map2(my_model, mynewdata, predict)) %>%
select(Species, mynewdata, my_new_pred) %>%
unnest(mynewdata, my_new_pred) %>%
rename(modeled = my_new_pred, measured = Sepal.Length) %>%
gather("Type", "Sepal.Length", modeled, measured)
嵌套的
predictions_tall对象看起来像这样:
predictions_tall %>% nest(-Species, -type) %>% as.tibble()
# A tibble: 6 x 3
Species type data
<fctr> <chr> <list>
1 setosa modeled <data.frame [32 x 4]>
2 versicolor modeled <data.frame [33 x 4]>
3 virginica modeled <data.frame [35 x 4]>
4 setosa measured <data.frame [32 x 4]>
5 versicolor measured <data.frame [33 x 4]>
6 virginica measured <data.frame [35 x 4]>
最后,展示预测结果的图表:
predictions_tall %>%
ggplot(aes(x = Petal.Length, y = Sepal.Length)) +
geom_line(aes(color = Species, linetype = Type))
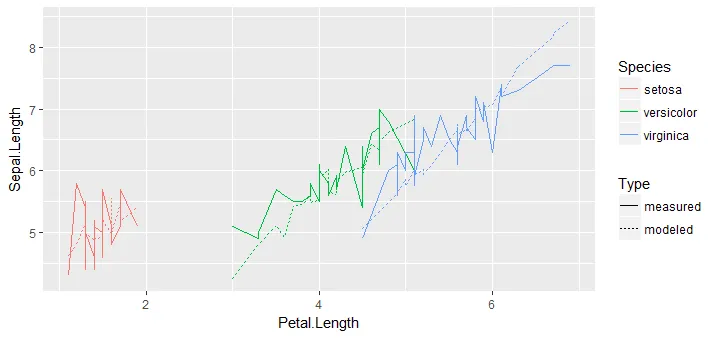
翻译 - the broom way
我现在已经更新,只使用每个组的模型计算预测。
这种方法使用broom包-具体来说是augment函数-添加拟合值。更多信息请参见此处:https://cran.r-project.org/web/packages/broom/vignettes/broom.html
由于您没有提供数据,在这里我使用iris。
library(tidyverse)
library(broom)
set.seed(1234)
myiris <- iris[sample(nrow(iris), replace = F),]
origiris <-
myiris[1:25,] %>%
nest(-Species) %>%
rename(origdata = data)
prediris <-
myiris[101:150,] %>%
nest(-Species) %>%
rename(preddata = data)
iris_mod <-
origiris %>%
mutate(mod = map(origdata, ~ MASS::rlm(Sepal.Length ~ Petal.Length + Petal.Width, data = .)))
首先为原始数据集获取拟合值(非必需,仅供说明):
# get fitted values for the first dataset (origdata)
origiris_aug <-
iris_mod %>%
mutate(origpred = map(mod, augment)) %>%
unnest(origpred) %>%
as.tibble()
原始的iris_aug预测数据框如下所示:
origiris_aug
Species .rownames Sepal.Length Petal.Length Petal.Width .fitted .se.fit .resid
<fctr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 setosa 18 5.1 1.4 0.3 5.002797 0.1514850 0.09720290
2 setosa 2 4.9 1.4 0.2 4.931824 0.1166911 -0.03182417
3 setosa 34 5.5 1.4 0.2 4.931824 0.1166911 0.56817583
4 setosa 40 5.1 1.5 0.2 4.981975 0.1095883 0.11802526
5 setosa 39 4.4 1.3 0.2 4.881674 0.1422123 -0.48167359
6 setosa 36 5.0 1.2 0.2 4.831523 0.1784156 0.16847698
7 setosa 25 4.8 1.9 0.2 5.182577 0.2357614 -0.38257703
8 setosa 31 4.8 1.6 0.2 5.032125 0.1241074 -0.23212531
9 setosa 42 4.5 1.3 0.3 4.952647 0.1760223 -0.45264653
10 setosa 21 5.4 1.7 0.2 5.082276 0.1542594 0.31772411
现在您实际想要的是对新数据集进行预测:
prediris_aug <-
iris_mod %>%
inner_join(prediris, by = "Species") %>%
map2_df(.x = iris_mod$mod, .y = prediris$preddata, .f = ~augment(.x, newdata = .y)) %>%
as.tibble()
prediris_aug 数据框如下所示:
prediris_aug
.rownames Sepal.Length Sepal.Width Petal.Length Petal.Width .fitted .se.fit
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 105 6.5 3.0 5.8 2.2 8.557908 3.570269
2 115 5.8 2.8 5.1 2.4 8.348800 3.666631
3 117 6.5 3.0 5.5 1.8 8.123565 3.005888
4 139 6.0 3.0 4.8 1.8 7.772511 2.812748
5 103 7.1 3.0 5.9 2.1 8.537086 3.475224
6 107 4.9 2.5 4.5 1.7 7.551086 2.611123
7 119 7.7 2.6 6.9 2.3 9.180537 4.000412
8 135 6.1 2.6 5.6 1.4 7.889823 2.611457
9 124 6.3 2.7 4.9 1.8 7.822661 2.838502
10 118 7.7 3.8 6.7 2.2 9.009263 3.825613