我目前正在开发一种算法,用于确定细菌聚集的(Brightfield)显微镜图像的质心位置。这是图像处理中一个重要的开放性问题。
这个问题是Python/OpenCV — Matching Centroid Points of Bacteria in Two Images的后续。
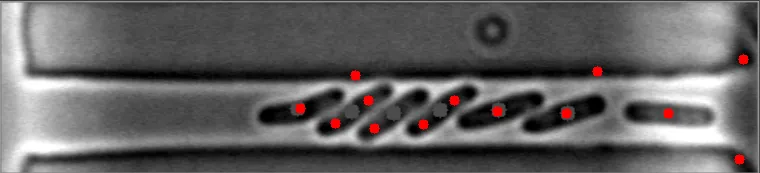
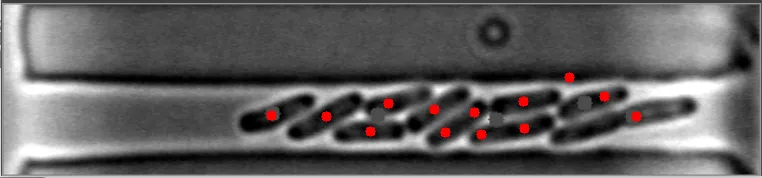
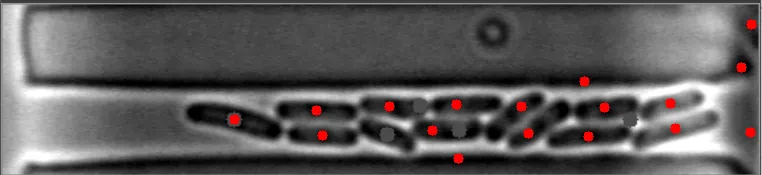
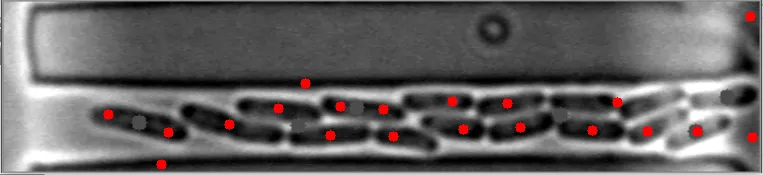
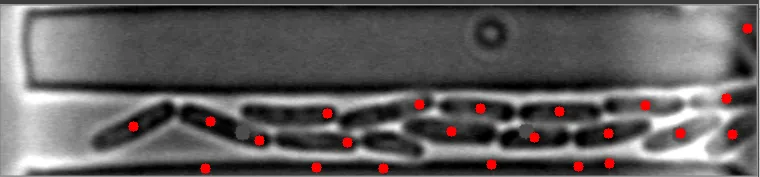
目前,该算法对于稀疏、分散的细菌非常有效。然而,当细菌聚集在一起时,它变得完全无效。
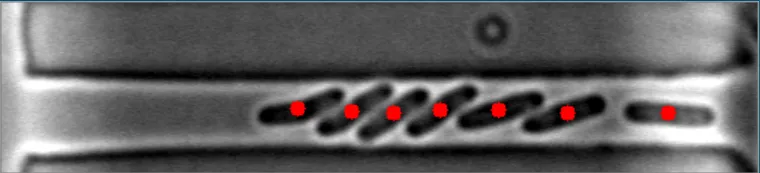
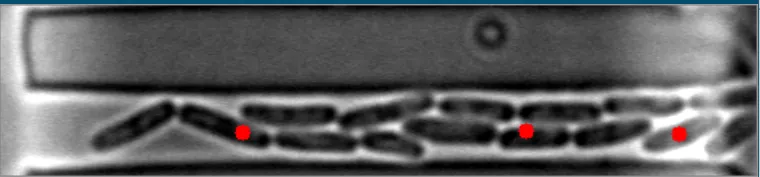
在这些图像中,注意到细菌的质心位置被有效地定位。 亮场图像#1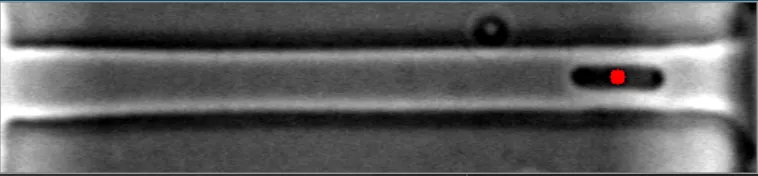 亮场图像#2
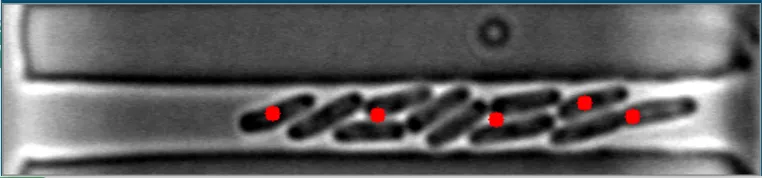
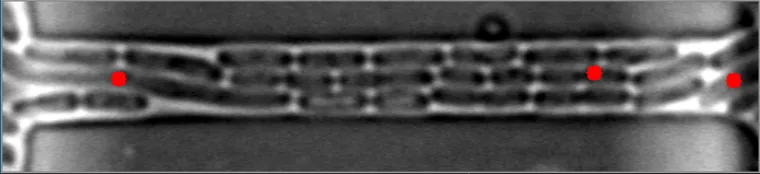
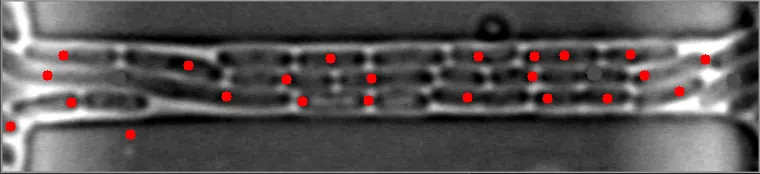
亮场图像#2
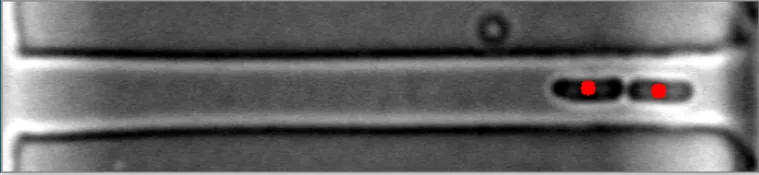
理想情况下,我希望至少能在发布的几幅图像中进行改进。
这个问题是Python/OpenCV — Matching Centroid Points of Bacteria in Two Images的后续。
目前,该算法对于稀疏、分散的细菌非常有效。然而,当细菌聚集在一起时,它变得完全无效。
在这些图像中,注意到细菌的质心位置被有效地定位。 亮场图像#1
 亮场图像#2
亮场图像#2

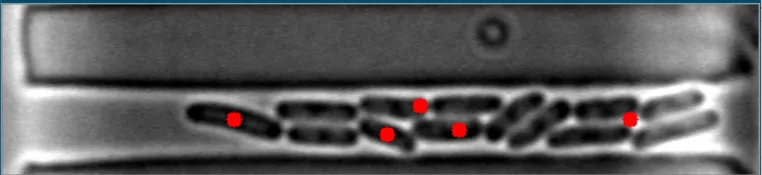
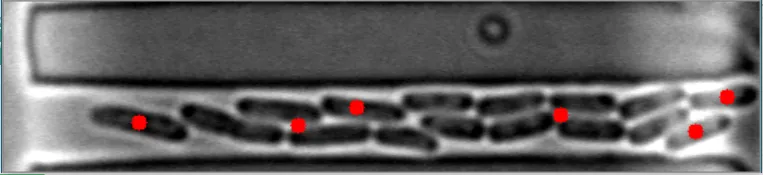
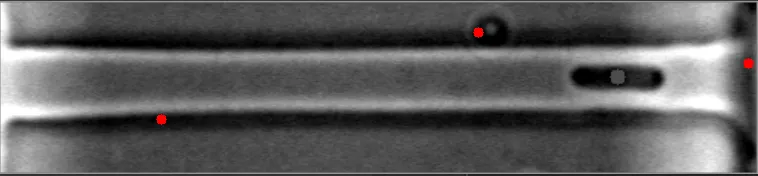
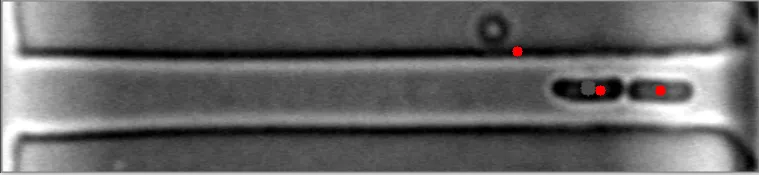
然而,当细菌在不同水平聚集时,算法会失败。
原始图像
我希望优化我的当前算法,使其对这些类型的图像更加强健。 这是我正在运行的程序。
import cv2
import numpy as np
import os
kernel = np.array([[0, 0, 1, 0, 0],
[0, 1, 1, 1, 0],
[1, 1, 1, 1, 1],
[0, 1, 1, 1, 0],
[0, 0, 1, 0, 0]], dtype=np.uint8)
def e_d(image, it):
image = cv2.erode(image, kernel, iterations=it)
image = cv2.dilate(image, kernel, iterations=it)
return image
path = r"(INSERT IMAGE DIRECTORY HERE)"
img_files = [file for file in os.listdir(path)]
def segment_index(index: int):
segment_file(img_files[index])
def segment_file(img_file: str):
img_path = path + "\\" + img_file
print(img_path)
img = cv2.imread(img_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# Applying adaptive mean thresholding
th = cv2.adaptiveThreshold(img, 255, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY_INV, 11, 2)
# Removing small noise
th = e_d(th.copy(), 1)
# Finding contours with RETR_EXTERNAL flag and removing undesired contours and
# drawing them on a new image.
cnt, hie = cv2.findContours(th, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
cntImg = th.copy()
for contour in cnt:
x, y, w, h = cv2.boundingRect(contour)
# Eliminating the contour if its width is more than half of image width
# (bacteria will not be that big).
if w > img.shape[1] / 2:
continue
cntImg = cv2.drawContours(cntImg, [cv2.convexHull(contour)], -1, 255, -1)
# Removing almost all the remaining noise.
# (Some big circular noise will remain along with bacteria contours)
cntImg = e_d(cntImg, 3)
# Finding new filtered contours again
cnt2, hie2 = cv2.findContours(cntImg, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_NONE)
# Now eliminating circular type noise contours by comparing each contour's
# extent of overlap with its enclosing circle.
finalContours = [] # This will contain the final bacteria contours
for contour in cnt2:
# Finding minimum enclosing circle
(x, y), radius = cv2.minEnclosingCircle(contour)
center = (int(x), int(y))
radius = int(radius)
# creating a image with only this circle drawn on it(filled with white colour)
circleImg = np.zeros(img.shape, dtype=np.uint8)
circleImg = cv2.circle(circleImg, center, radius, 255, -1)
# creating a image with only the contour drawn on it(filled with white colour)
contourImg = np.zeros(img.shape, dtype=np.uint8)
contourImg = cv2.drawContours(contourImg, [contour], -1, 255, -1)
# White pixels not common in both contour and circle will remain white
# else will become black.
union_inter = cv2.bitwise_xor(circleImg, contourImg)
# Finding ratio of the extent of overlap of contour to its enclosing circle.
# Smaller the ratio, more circular the contour.
ratio = np.sum(union_inter == 255) / np.sum(circleImg == 255)
# Storing only non circular contours(bacteria)
if ratio > 0.55:
finalContours.append(contour)
finalContours = np.asarray(finalContours)
# Finding center of bacteria and showing it.
bacteriaImg = cv2.cvtColor(img, cv2.COLOR_GRAY2BGR)
for bacteria in finalContours:
M = cv2.moments(bacteria)
cx = int(M['m10'] / M['m00'])
cy = int(M['m01'] / M['m00'])
bacteriaImg = cv2.circle(bacteriaImg, (cx, cy), 5, (0, 0, 255), -1)
cv2.imshow("bacteriaImg", bacteriaImg)
cv2.waitKey(0)
# Segment Each Image
for i in range(len(img_files)):
segment_index(i)
理想情况下,我希望至少能在发布的几幅图像中进行改进。