我有一个geom_gene_arrow图表,我需要使用ggrepel来防止标签重叠。不幸的是,我无法让它正常工作,并且会出现错误,提示“找不到函数”geom_text_repel“'
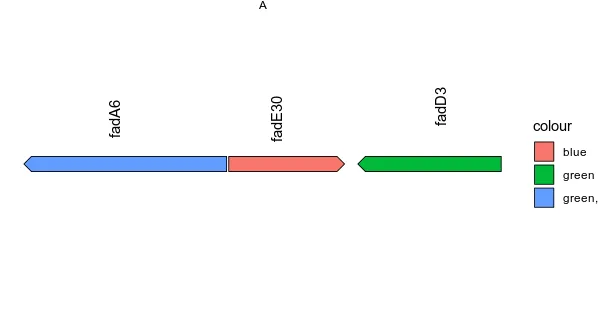
只使用geom_text的可行示例:
> ggplot(data, aes(xmin = start, xmax = end, y = genome, fill = colour, forward = direction)) +
geom_gene_arrow() +
geom_text(aes(x = end - ((end-start)/2), y = 1.2, label = gene, angle=90)) +
facet_wrap(~ genome, scales = "free", ncol = 1) +
theme_void()+
xlab("")
随着引入geom_text_repel,它失败了。
> ggplot(data, aes(xmin = start, xmax = end, y = genome, fill = colour, forward = direction)) +
geom_gene_arrow() +
geom_text_repel(aes(x = end - ((end-start)/2), y = 1.2, label = gene, angle=90)) +
facet_wrap(~ genome, scales = "free", ncol = 1) +
theme_void()+
xlab("")
示例数据:
genome start end gene function colour direction
A 11638 12786 fadA6 ringdegradation green, -1
A 12798 13454 fadE30 cleavage, blue 1
A 13529 14341 fadD3 ringdegradation green -1
任何关于我做错的事情的洞察都非常感激!
ggrepel?此外,在你的ggplot代码中,提到了start、end和genome变量,但这些变量在你的示例中并不存在。你能否提供一个可重现的数据集示例?(请按照此教程操作:https://dev59.com/eG025IYBdhLWcg3whGSx) - dc37ggrepel时,你没有注意到installation of package ‘ggrepel'had non-zero exit status的错误吗?一个函数未找到的常见错误是未加载包(例如在运行代码之前执行library(ggrepel))。 - dc37