也许有人可以帮我。我花了好几天的时间,但无法解决这个问题。谢谢您提前的帮助。
我想要将两个洛仑兹函数拟合到我的实验数据中。我将我的方程简化为两个洛仑兹函数lorentz1和lorentz2。然后,我定义了另外两个函数L1和L2,只需将常数cnst乘以它们。我有所有需要拟合的四个参数: cnst1、cnst2、tau1、tau2。
我使用lmfit:Model and minimize(可能都使用同一种方法)。
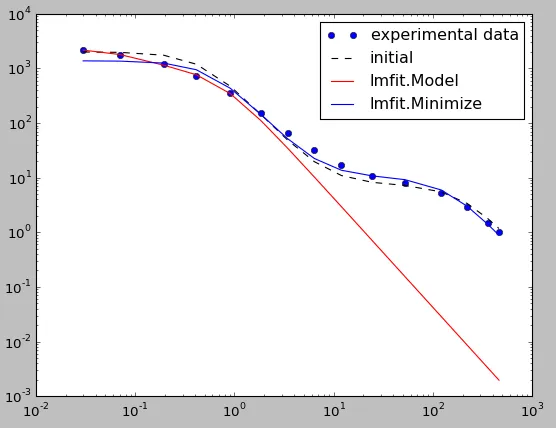
初始拟合参数设置得更接近一个良好的拟合结果,但使用lmfit进行最小化时会失控(如下面第一个图所示):
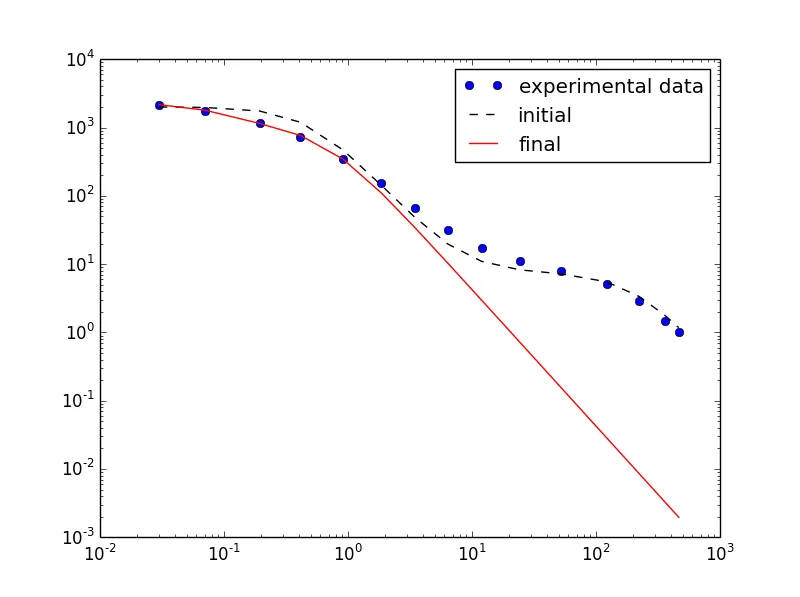
另一方面,将参数限制在非常接近初始值的范围内(强制为初始值):
使用以下参数: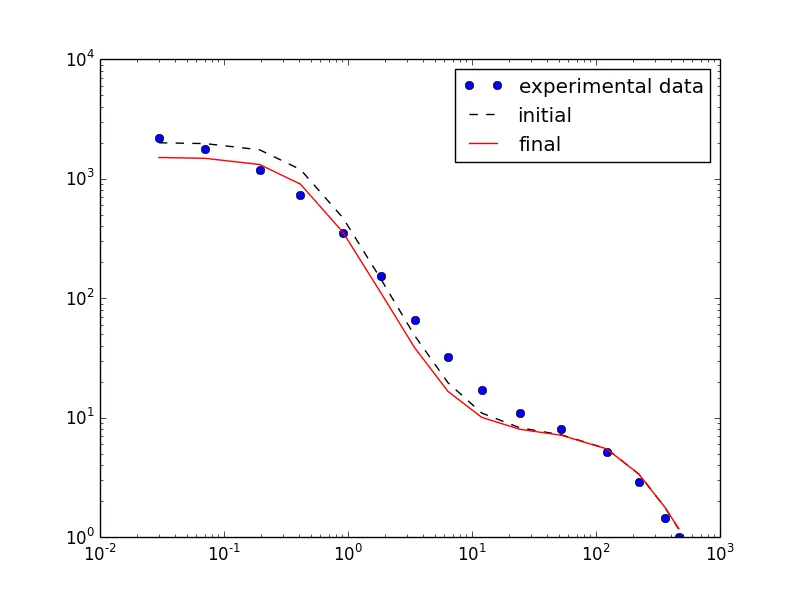
我想要将两个洛仑兹函数拟合到我的实验数据中。我将我的方程简化为两个洛仑兹函数lorentz1和lorentz2。然后,我定义了另外两个函数L1和L2,只需将常数cnst乘以它们。我有所有需要拟合的四个参数: cnst1、cnst2、tau1、tau2。
我使用lmfit:Model and minimize(可能都使用同一种方法)。
初始拟合参数设置得更接近一个良好的拟合结果,但使用lmfit进行最小化时会失控(如下面第一个图所示):
使用这些参数:
params.add('cnst1', value=1e3 , min=1e2, max=1e5)
params.add('cnst2', value=3e5, min=1e2, max=1e6)
params.add('tau1', value=2e0, min=0, max=1e2)
params.add('tau2', value=5e-3, min=0, max=10)
但错误率很低:
cnst1: 117.459806 +/- 14.67188 (12.49%) (init= 1000)
cnst2: 413.959032 +/- 44.21042 (10.68%) (init= 300000)
tau1: 11.0343531 +/- 1.065570 (9.66%) (init= 2)
tau2: 1.55259664 +/- 0.125853 (8.11%) (init= 0.005)
另一方面,将参数限制在非常接近初始值的范围内(强制为初始值):
使用以下参数:

#params.add('cnst1', value=1e3 , min=0.1e3, max=1e3)
#params.add('cnst2', value=3e5, min=1e3, max=1e6)
#params.add('tau1', value=2e0, min=0, max=2)
#params.add('tau2', value=5e-3, min=0, max=10)
虽然 fit 看起来更好,但误差值非常大:
[[Variables]]
cnst1: 752.988629 +/- 221.3098 (29.39%) (init= 1000)
cnst2: 3.0159e+05 +/- 3.05e+07 (10113.40%) (init= 300000)
tau1: 1.99684317 +/- 0.600748 (30.08%) (init= 2)
tau2: 0.00497806 +/- 0.289651 (5818.56%) (init= 0.005)
这是完整的代码:
import numpy as np
from lmfit import Model, minimize, Parameters, report_fit
import matplotlib.pyplot as plt
x = np.array([0.02988, 0.07057,0.19365,0.4137,0.91078,1.85075,3.44353,6.39428,\
11.99302,24.37024,52.58804,121.71927,221.53799,358.27392,464.70405])
y = 1.0 / np.array([4.60362E-4,5.63559E-4,8.44538E-4,0.00138,0.00287,0.00657,0.01506,\
0.03119,0.0584,0.09153,0.12538,0.19389,0.34391,0.68869,1.0])
def lorentz1(x, tau):
L = tau / ( 1 + (x*tau)**2 )
return(L)
def lorentz2(x, tau):
L = tau**2 / ( 1 + (x*tau)**2 )
return(L)
def L1(x,cnst1,tau1):
L1 = cnst1 * lorentz1(x,tau1)
return (L1)
def L2(x, cnst2, tau2):
L2 = cnst2 * lorentz2(x,tau2)
return (L2)
def L_min(params, x, y):
cnst1 = params['cnst1'].value
cnst2 = params['cnst2'].value
tau1 = params['tau1'].value
tau2 = params['tau2'].value
L_total = L1(x, cnst1, tau1) + L2(x, cnst2, tau2)
resids = L_total - y
return resids
#params = mod.make_params( cnst1=10e2, cnst2=3e5, tau1=2e0, tau2=0.5e-2)
params = Parameters()
#params.add('cnst1', value=1e3 , min=0.1e3, max=1e3)
#params.add('cnst2', value=3e5, min=1e3, max=1e6)
#params.add('tau1', value=2e0, min=0, max=2)
#params.add('tau2', value=5e-3, min=0, max=10)
params.add('cnst1', value=1e3 , min=1e2, max=1e5)
params.add('cnst2', value=3e5, min=1e2, max=1e6)
params.add('tau1', value=2e0, min=0, max=1e2)
params.add('tau2', value=5e-3, min=0, max=10)
#1-----Model--------------------
mod = Model(L1) + Model(L2)
result_mod = mod.fit(y, params, x=x)
print('---results from lmfit.Model----')
print(result_mod.fit_report())
#2---minimize-----------
result_min = minimize(L_min, params, args=(x,y))
final_min = y + result_min.residual
print('---results from lmfit.minimize----')
report_fit(params)
#-------Plot------
plt.close('all')
plt.loglog(x, y,'bo' , label='experimental data')
plt.loglog(x, result_mod.init_fit, 'k--', label='initial')
plt.loglog(x, result_mod.best_fit, 'r-', label='final')
plt.legend()
plt.show()