以下代码大量摘自一个sparklyr博客文章,仅用于提供示例:
library(sparklyr)
library(dplyr)
# If needed, install Spark locally via `spark_install()`
sc <- spark_connect(master = "local")
iris_tbl <- copy_to(sc, iris)
# split the data into train and validation sets
iris_data <- iris_tbl %>%
sdf_partition(train = 2/3, validation = 1/3, seed = 123)
iris_pipeline <- ml_pipeline(sc) %>%
ft_dplyr_transformer(
iris_data$train %>%
mutate(Sepal_Length = log(Sepal_Length),
Sepal_Width = Sepal_Width ^ 2)
) %>%
ft_string_indexer("Species", "label")
iris_pipeline_model <- iris_pipeline %>%
ml_fit(iris_data$train)
iris_vector_assembler <- ft_vector_assembler(
sc,
input_cols = setdiff(colnames(iris_data$train), "Species"),
output_col = "features"
)
random_forest <- ml_random_forest_classifier(sc,features_col = "features")
# obtain the labels from the fitted StringIndexerModel
iris_labels <- iris_pipeline_model %>%
ml_stage("string_indexer") %>%
ml_labels()
# IndexToString will convert the predicted numeric values back to class labels
iris_index_to_string <- ft_index_to_string(sc, "prediction", "predicted_label",
labels = iris_labels)
# construct a pipeline with these stages
iris_prediction_pipeline <- ml_pipeline(
iris_pipeline, # pipeline from previous section
iris_vector_assembler,
random_forest,
iris_index_to_string
)
# fit to data and make some predictions
iris_prediction_model <- iris_prediction_pipeline %>%
ml_fit(iris_data$train)
iris_predictions <- iris_prediction_model %>%
ml_transform(iris_data$validation)
iris_predictions %>%
select(Species, label:predicted_label) %>%
glimpse()
在参考这里的建议后,经过反复试验,我成功地将底层决策树制成字符串形式的 "if/else" 格式并打印了出来:
model_stage <- iris_prediction_model$stages[[3]]
spark_jobj(model_stage) %>% invoke(., "toDebugString") %>% cat()
##print out below##
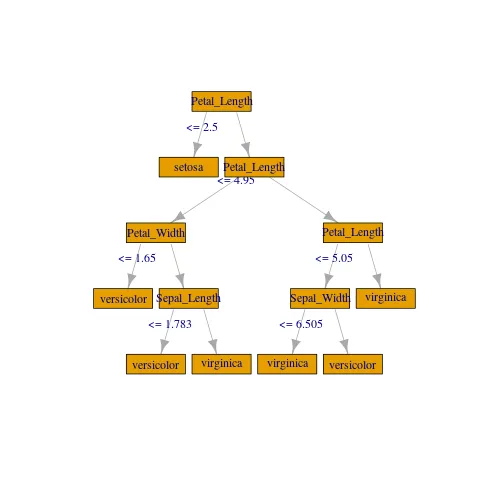
RandomForestClassificationModel (uid=random_forest_classifier_5c6a1934c8e) with 20 trees
Tree 0 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 4.95)
If (feature 3 <= 1.65)
Predict: 0.0
Else (feature 3 > 1.65)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Else (feature 2 > 4.95)
If (feature 2 <= 5.05)
If (feature 1 <= 6.505000000000001)
Predict: 2.0
Else (feature 1 > 6.505000000000001)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 1 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.75)
If (feature 1 <= 5.0649999999999995)
If (feature 3 <= 1.05)
Predict: 0.0
Else (feature 3 > 1.05)
If (feature 0 <= 1.8000241202036602)
Predict: 2.0
Else (feature 0 > 1.8000241202036602)
Predict: 0.0
Else (feature 1 > 5.0649999999999995)
If (feature 0 <= 1.8000241202036602)
Predict: 0.0
Else (feature 0 > 1.8000241202036602)
If (feature 2 <= 5.05)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 2 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 0 <= 1.7664051342320237)
Predict: 0.0
Else (feature 0 > 1.7664051342320237)
If (feature 3 <= 1.45)
If (feature 2 <= 4.85)
Predict: 0.0
Else (feature 2 > 4.85)
Predict: 2.0
Else (feature 3 > 1.45)
If (feature 3 <= 1.65)
If (feature 1 <= 8.125)
Predict: 2.0
Else (feature 1 > 8.125)
Predict: 0.0
Else (feature 3 > 1.65)
Predict: 2.0
Tree 3 (weight 1.0):
If (feature 0 <= 1.6675287895788053)
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
Predict: 0.0
Else (feature 0 > 1.6675287895788053)
If (feature 3 <= 1.75)
If (feature 3 <= 1.55)
If (feature 1 <= 7.025)
If (feature 2 <= 4.55)
Predict: 0.0
Else (feature 2 > 4.55)
Predict: 2.0
Else (feature 1 > 7.025)
Predict: 0.0
Else (feature 3 > 1.55)
If (feature 2 <= 5.05)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 4 (weight 1.0):
If (feature 2 <= 4.85)
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
Predict: 0.0
Else (feature 2 > 4.85)
If (feature 2 <= 5.05)
If (feature 0 <= 1.8484238118815566)
Predict: 2.0
Else (feature 0 > 1.8484238118815566)
Predict: 0.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 5 (weight 1.0):
If (feature 2 <= 1.65)
Predict: 1.0
Else (feature 2 > 1.65)
If (feature 3 <= 1.65)
If (feature 0 <= 1.8325494627242664)
Predict: 0.0
Else (feature 0 > 1.8325494627242664)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Else (feature 3 > 1.65)
Predict: 2.0
Tree 6 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 5.05)
If (feature 3 <= 1.75)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 7 (weight 1.0):
If (feature 3 <= 0.55)
Predict: 1.0
Else (feature 3 > 0.55)
If (feature 3 <= 1.65)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
Predict: 2.0
Else (feature 3 > 1.65)
If (feature 2 <= 4.85)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Else (feature 2 > 4.85)
Predict: 2.0
Tree 8 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.85)
If (feature 2 <= 4.85)
Predict: 0.0
Else (feature 2 > 4.85)
If (feature 0 <= 1.8794359129669855)
Predict: 2.0
Else (feature 0 > 1.8794359129669855)
If (feature 3 <= 1.55)
Predict: 0.0
Else (feature 3 > 1.55)
Predict: 0.0
Else (feature 3 > 1.85)
Predict: 2.0
Tree 9 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Tree 10 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
If (feature 2 <= 5.05)
If (feature 3 <= 1.55)
Predict: 2.0
Else (feature 3 > 1.55)
If (feature 3 <= 1.75)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 11 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 2 <= 5.05)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 3 <= 1.75)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Else (feature 2 > 5.05)
Predict: 2.0
Tree 12 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.75)
If (feature 3 <= 1.35)
Predict: 0.0
Else (feature 3 > 1.35)
If (feature 0 <= 1.695573522904327)
Predict: 0.0
Else (feature 0 > 1.695573522904327)
If (feature 1 <= 8.125)
Predict: 2.0
Else (feature 1 > 8.125)
Predict: 0.0
Else (feature 3 > 1.75)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Tree 13 (weight 1.0):
If (feature 3 <= 0.55)
Predict: 1.0
Else (feature 3 > 0.55)
If (feature 2 <= 4.95)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 0 <= 1.8000241202036602)
If (feature 1 <= 9.305)
Predict: 2.0
Else (feature 1 > 9.305)
Predict: 0.0
Else (feature 0 > 1.8000241202036602)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Tree 14 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 3 <= 1.65)
If (feature 3 <= 1.45)
Predict: 0.0
Else (feature 3 > 1.45)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
Else (feature 3 > 1.65)
If (feature 0 <= 1.7833559100698644)
If (feature 0 <= 1.7664051342320237)
Predict: 2.0
Else (feature 0 > 1.7664051342320237)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Tree 15 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 3 <= 1.75)
If (feature 2 <= 4.95)
Predict: 0.0
Else (feature 2 > 4.95)
If (feature 1 <= 8.125)
Predict: 2.0
Else (feature 1 > 8.125)
If (feature 0 <= 1.9095150692894909)
Predict: 0.0
Else (feature 0 > 1.9095150692894909)
Predict: 2.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 16 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 0 <= 1.7491620461964392)
Predict: 0.0
Else (feature 0 > 1.7491620461964392)
If (feature 3 <= 1.75)
If (feature 2 <= 4.75)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 0 <= 1.8164190316151556)
Predict: 2.0
Else (feature 0 > 1.8164190316151556)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 17 (weight 1.0):
If (feature 0 <= 1.695573522904327)
If (feature 2 <= 1.65)
Predict: 1.0
Else (feature 2 > 1.65)
Predict: 0.0
Else (feature 0 > 1.695573522904327)
If (feature 2 <= 4.75)
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
Predict: 0.0
Else (feature 2 > 4.75)
If (feature 3 <= 1.75)
If (feature 1 <= 5.0649999999999995)
Predict: 2.0
Else (feature 1 > 5.0649999999999995)
If (feature 3 <= 1.65)
Predict: 0.0
Else (feature 3 > 1.65)
Predict: 0.0
Else (feature 3 > 1.75)
Predict: 2.0
Tree 18 (weight 1.0):
If (feature 3 <= 0.8)
Predict: 1.0
Else (feature 3 > 0.8)
If (feature 3 <= 1.65)
Predict: 0.0
Else (feature 3 > 1.65)
If (feature 0 <= 1.7833559100698644)
Predict: 0.0
Else (feature 0 > 1.7833559100698644)
Predict: 2.0
Tree 19 (weight 1.0):
If (feature 2 <= 2.5)
Predict: 1.0
Else (feature 2 > 2.5)
If (feature 2 <= 4.95)
If (feature 1 <= 8.705)
Predict: 0.0
Else (feature 1 > 8.705)
If (feature 2 <= 4.85)
Predict: 0.0
Else (feature 2 > 4.85)
If (feature 0 <= 1.8164190316151556)
Predict: 2.0
Else (feature 0 > 1.8164190316151556)
Predict: 0.0
Else (feature 2 > 4.95)
Predict: 2.0
如您所见,这种格式不太适合传入我所见过的许多美丽的决策树图形可视化方法之一(例如Revolution Analytics或StatMethods)。
V(g)$color <- ...来设置节点的颜色。 - zero323