我了解到所需的森林图是在模型公式中简单跳过分层的RX变量。如果是这样,我们可以在内部插入一个if语句,忽略与数据列名称不完全对应的公式部分(例如,"strata(rx)"不是列名)。
方法1
如果你足够熟悉R以修改函数,请运行trace(ggforest, edit = TRUE)并在弹出窗口中用以下版本替换allTerms <- lapply(...)(大约在第10-25行附近):
allTerms <- lapply(seq_along(terms), function(i) {
var <- names(terms)[i]
if(var %in% colnames(data)) {
if (terms[i] %in% c("factor", "character")) {
adf <- as.data.frame(table(data[, var]))
cbind(var = var, adf, pos = 1:nrow(adf))
}
else if (terms[i] == "numeric") {
data.frame(var = var, Var1 = "", Freq = nrow(data),
pos = 1)
}
else {
vars = grep(paste0("^", var, "*."), coef$term,
value = TRUE)
data.frame(var = vars, Var1 = "", Freq = nrow(data),
pos = seq_along(vars))
}
} else {
message(var, "is not found in data columns, and will be skipped.")
}
})
ggforest(stratamodel)

这种修改不会保留到后续的R会话中。如果您想在当前会话中撤消修改,只需运行untrace(ggforest)。
方法2
如果您希望永久修改函数以备将来使用/不想对库函数进行操作,请保存以下函数:
ggforest2 <- function (model, data = NULL, main = "Hazard ratio",
cpositions = c(0.02, 0.22, 0.4), fontsize = 0.7,
refLabel = "reference", noDigits = 2) {
conf.high <- conf.low <- estimate <- NULL
stopifnot(class(model) == "coxph")
data <- survminer:::.get_data(model, data = data)
terms <- attr(model$terms, "dataClasses")[-1]
coef <- as.data.frame(broom::tidy(model))
gmodel <- broom::glance(model)
allTerms <- lapply(seq_along(terms), function(i) {
var <- names(terms)[i]
if(var %in% colnames(data)) {
if (terms[i] %in% c("factor", "character")) {
adf <- as.data.frame(table(data[, var]))
cbind(var = var, adf, pos = 1:nrow(adf))
}
else if (terms[i] == "numeric") {
data.frame(var = var, Var1 = "", Freq = nrow(data),
pos = 1)
}
else {
vars = grep(paste0("^", var, "*."), coef$term,
value = TRUE)
data.frame(var = vars, Var1 = "", Freq = nrow(data),
pos = seq_along(vars))
}
} else {
message(var, "is not found in data columns, and will be skipped.")
}
})
allTermsDF <- do.call(rbind, allTerms)
colnames(allTermsDF) <- c("var", "level", "N",
"pos")
inds <- apply(allTermsDF[, 1:2], 1, paste0, collapse = "")
rownames(coef) <- gsub(coef$term, pattern = "`", replacement = "")
toShow <- cbind(allTermsDF, coef[inds, ])[, c("var", "level", "N", "p.value",
"estimate", "conf.low",
"conf.high", "pos")]
toShowExp <- toShow[, 5:7]
toShowExp[is.na(toShowExp)] <- 0
toShowExp <- format(exp(toShowExp), digits = noDigits)
toShowExpClean <- data.frame(toShow, pvalue = signif(toShow[, 4], noDigits + 1),
toShowExp)
toShowExpClean$stars <- paste0(round(toShowExpClean$p.value, noDigits + 1), " ",
ifelse(toShowExpClean$p.value < 0.05, "*", ""),
ifelse(toShowExpClean$p.value < 0.01, "*", ""),
ifelse(toShowExpClean$p.value < 0.001, "*", ""))
toShowExpClean$ci <- paste0("(", toShowExpClean[, "conf.low.1"],
" - ", toShowExpClean[, "conf.high.1"], ")")
toShowExpClean$estimate.1[is.na(toShowExpClean$estimate)] = refLabel
toShowExpClean$stars[which(toShowExpClean$p.value < 0.001)] = "<0.001 ***"
toShowExpClean$stars[is.na(toShowExpClean$estimate)] = ""
toShowExpClean$ci[is.na(toShowExpClean$estimate)] = ""
toShowExpClean$estimate[is.na(toShowExpClean$estimate)] = 0
toShowExpClean$var = as.character(toShowExpClean$var)
toShowExpClean$var[duplicated(toShowExpClean$var)] = ""
toShowExpClean$N <- paste0("(N=", toShowExpClean$N, ")")
toShowExpClean <- toShowExpClean[nrow(toShowExpClean):1, ]
rangeb <- range(toShowExpClean$conf.low, toShowExpClean$conf.high,
na.rm = TRUE)
breaks <- axisTicks(rangeb/2, log = TRUE, nint = 7)
rangeplot <- rangeb
rangeplot[1] <- rangeplot[1] - diff(rangeb)
rangeplot[2] <- rangeplot[2] + 0.15 * diff(rangeb)
width <- diff(rangeplot)
y_variable <- rangeplot[1] + cpositions[1] * width
y_nlevel <- rangeplot[1] + cpositions[2] * width
y_cistring <- rangeplot[1] + cpositions[3] * width
y_stars <- rangeb[2]
x_annotate <- seq_len(nrow(toShowExpClean))
annot_size_mm <- fontsize *
as.numeric(grid::convertX(unit(theme_get()$text$size, "pt"), "mm"))
p <- ggplot(toShowExpClean,
aes(seq_along(var), exp(estimate))) +
geom_rect(aes(xmin = seq_along(var) - 0.5,
xmax = seq_along(var) + 0.5,
ymin = exp(rangeplot[1]),
ymax = exp(rangeplot[2]),
fill = ordered(seq_along(var)%%2 + 1))) +
scale_fill_manual(values = c("#FFFFFF33", "#00000033"), guide = "none") +
geom_point(pch = 15, size = 4) +
geom_errorbar(aes(ymin = exp(conf.low), ymax = exp(conf.high)),
width = 0.15) +
geom_hline(yintercept = 1, linetype = 3) +
coord_flip(ylim = exp(rangeplot)) +
ggtitle(main) +
scale_y_log10(name = "", labels = sprintf("%g", breaks),
expand = c(0.02, 0.02), breaks = breaks) +
theme_light() +
theme(panel.grid.minor.y = element_blank(),
panel.grid.minor.x = element_blank(),
panel.grid.major.y = element_blank(),
legend.position = "none",
panel.border = element_blank(),
axis.title.y = element_blank(),
axis.text.y = element_blank(),
axis.ticks.y = element_blank(),
plot.title = element_text(hjust = 0.5)) +
xlab("") +
annotate(geom = "text", x = x_annotate, y = exp(y_variable),
label = toShowExpClean$var, fontface = "bold",
hjust = 0, size = annot_size_mm) +
annotate(geom = "text", x = x_annotate, y = exp(y_nlevel), hjust = 0,
label = toShowExpClean$level,
vjust = -0.1, size = annot_size_mm) +
annotate(geom = "text", x = x_annotate, y = exp(y_nlevel),
label = toShowExpClean$N, fontface = "italic", hjust = 0,
vjust = ifelse(toShowExpClean$level == "", 0.5, 1.1),
size = annot_size_mm) +
annotate(geom = "text", x = x_annotate, y = exp(y_cistring),
label = toShowExpClean$estimate.1, size = annot_size_mm,
vjust = ifelse(toShowExpClean$estimate.1 == "reference", 0.5, -0.1)) +
annotate(geom = "text", x = x_annotate, y = exp(y_cistring),
label = toShowExpClean$ci, size = annot_size_mm,
vjust = 1.1, fontface = "italic") +
annotate(geom = "text", x = x_annotate, y = exp(y_stars),
label = toShowExpClean$stars, size = annot_size_mm,
hjust = -0.2, fontface = "italic") +
annotate(geom = "text", x = 0.5, y = exp(y_variable),
label = paste0("# Events: ", gmodel$nevent,
"; Global p-value (Log-Rank): ",
format.pval(gmodel$p.value.log, eps = ".001"),
" \nAIC: ", round(gmodel$AIC, 2),
"; Concordance Index: ", round(gmodel$concordance, 2)),
size = annot_size_mm, hjust = 0, vjust = 1.2, fontface = "italic")
gt <- ggplot_gtable(ggplot_build(p))
gt$layout$clip[gt$layout$name == "panel"] <- "off"
ggpubr::as_ggplot(gt)
}
这是目前ggforest函数的快照,包含了上述相同的修改。如果软件包的创建者在未来进行修改,可能会使此方法失效或过时。但目前情况下,ggforest2(stratamodel)将产生与第一种方法相同的结果。
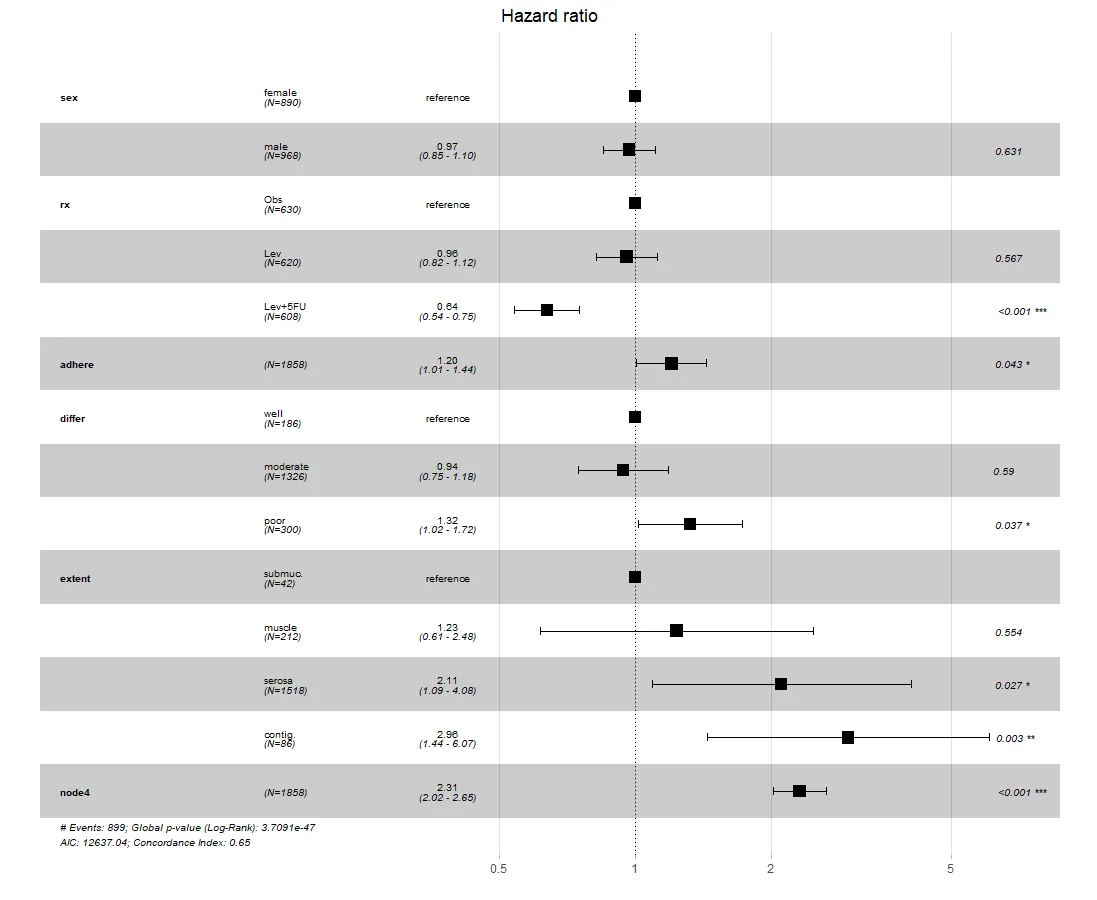

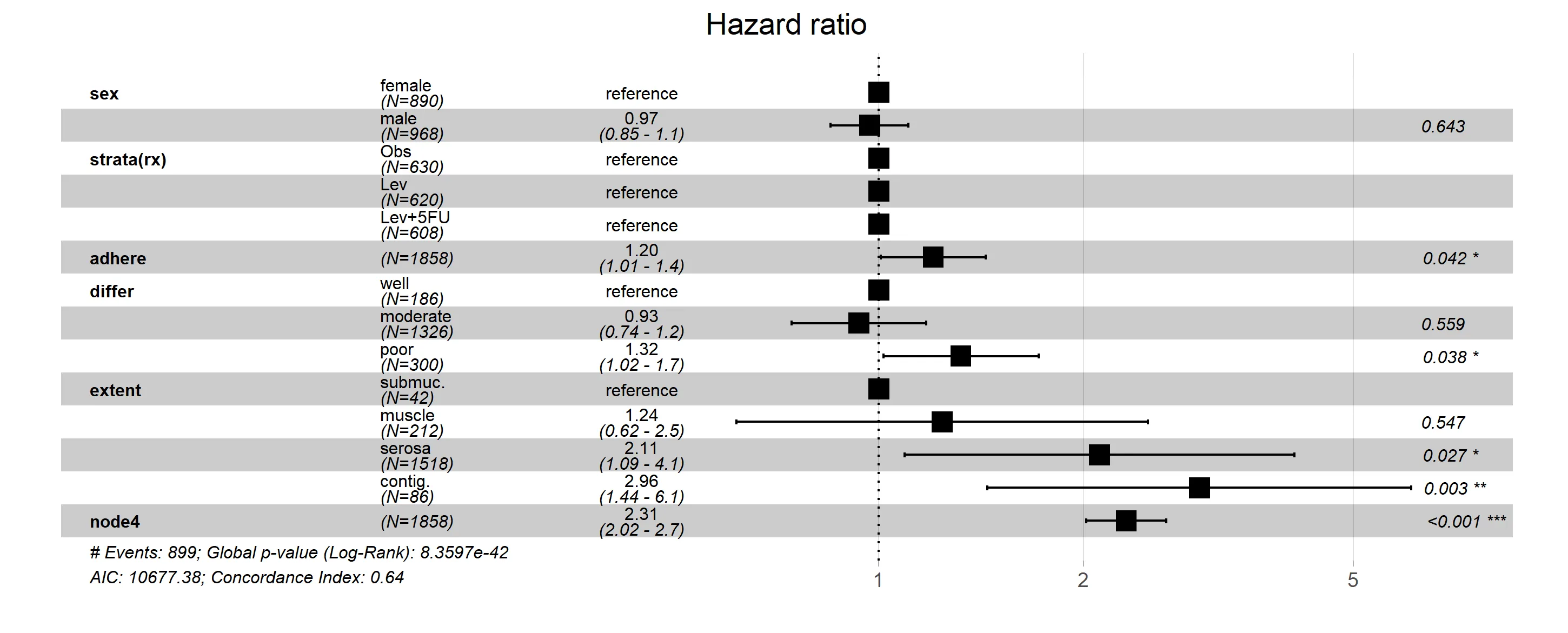
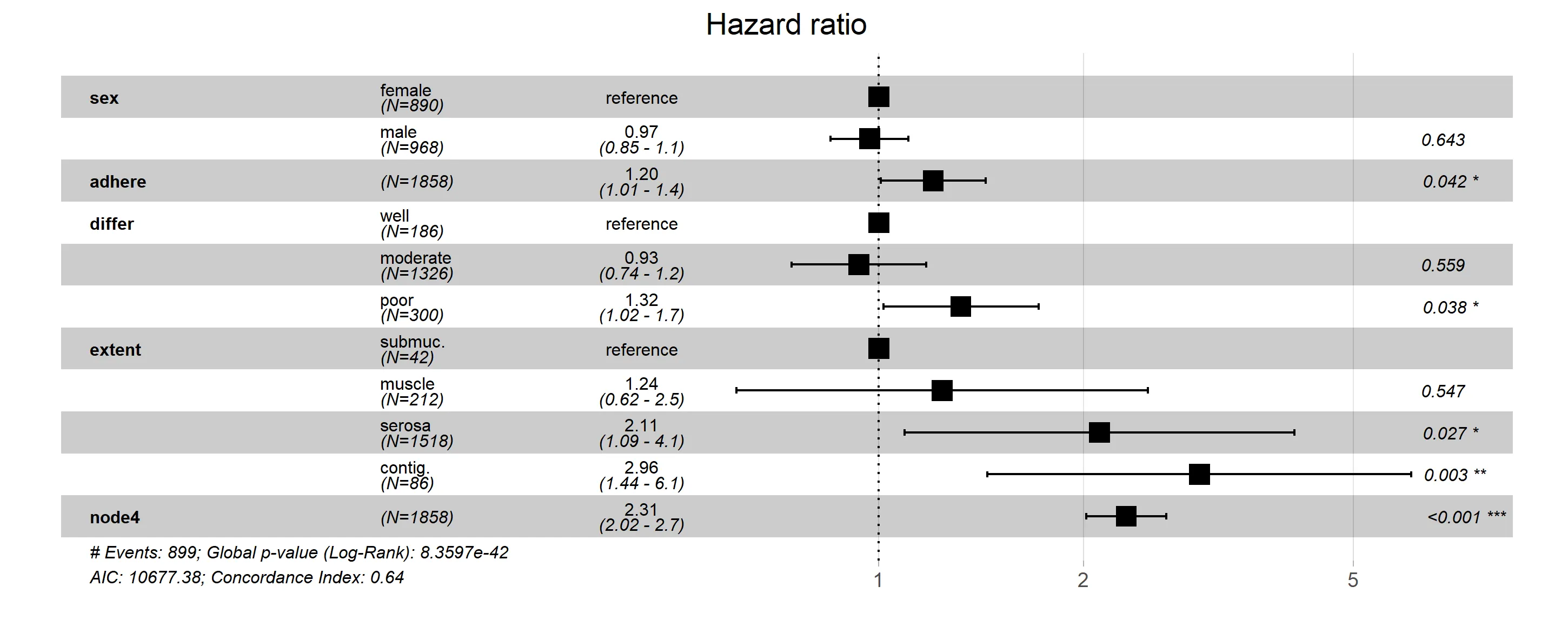
ggforest是一个包装器,因此使用底层的ggplot编码肯定可以工作。问题在于,熟悉分层模型和ggplot2的SO用户池可能不是很大。如果您可以包括分层模型图应该是什么样子的草图,那么您获得帮助的机会可能会增加。 - Z.Lin