我在ggplot2中使用了stat_smooth函数,决定想要“拟合优度”,于是用mgcv GAM来实现。我突然想到应该检查一下它们是否是相同的模型(stat_smooth vs mgcv's gam),于是我使用下面的代码进行了检查。
看起来,它们有不同的结果,如图所示( 图表:stat_smoother gam (红色),mgcv gam (黑色))。然而,我不知道它们为什么会有不同的结果。这是因为两者之间的某些默认参数不同吗?还是因为gam正在一个数字x上运行,而stat_smooth正在一个POSIXct x上运行(如果是这样的话-我不知道该怎么办)。看起来stat_smooth更平滑,但k值是相同的...
ggplot2中绘制gam输出的文章,但我真的想知道为什么stat_smooth和mgcv首先会给出不同的结果。我非常新手GAM(和R),所以很可能我错过了一些简单的东西。但是,在提问之前,我确实进行了谷歌搜索和在这个论坛上搜索。我的数据有点大,不容易共享,所以我使用了一个样本数据集 - 我把源代码放在了代码中,还有一个
dput()放在所有内容下面,然后是我的sessionInfo()。我试图提出一个高质量的问题,但这只是我的第二个问题。所以,欢迎提出建设性的批评。
谢谢!
library(readxl)
library(data.table)
library(ggplot2)
library(scales)
library(mgcv)
stackOF_data <- read_excel("mean-daily-flow-cumecs-vatnsdals.xlsx", sheet = "Data")
stackOF_data <- data.table(stackOF_data)
stackOF_data <- stackOF_data[,.(timeseries=as.POSIXct(Date,format("%Y-%m-%d")),mdf)]
a <- stackOF_data[,.(x=as.numeric(timeseries),y=mdf)]
a1 <- gam(y~s(x, k=100, bs="cs"),data=a)
a2=data.table(gam_mdf= predict(a1,a))
a2=cbind(timeseries=stackOF_data$timeseries,a2)
# see if predict and actual are the same
p <- ggplot() +
geom_line(data = a2, aes(x = timeseries, y = gam_mdf), size=1)+
scale_color_manual(values=c("black","magenta"))+
scale_y_continuous()+
scale_x_datetime(labels = date_format("%Y-%m-%d"), breaks = "1 month", minor_breaks = "1 week")+
theme(axis.text.x=element_text(angle=50, size=10,hjust=1))+
stat_smooth(data = stackOF_data, aes(x = (timeseries), y = mdf),method="gam", formula=y~s(x,k=100, bs="cs"), col="red", se=FALSE, size=1)
p
# data from: https://datamarket.com/data/set/235m/mean-daily-flow-cumecs-vatnsdalsa-river-1-jan-1972-31-dec-1974#!ds=235m&display=line&s=14l
> dput(a)
structure(list(x = c(126230400, 126316800, 126403200, 126489600,
126576000, 126662400, 126748800, 126835200, 126921600, 127008000,
127094400, 127180800, 127267200, 127353600, 127440000, 127526400,
127612800, 127699200, 127785600, 127872000, 127958400, 128044800,
128131200, 128217600, 128304000, 128390400, 128476800, 128563200,
128649600, 128736000, 128822400, 128908800, 128995200, 129081600,
129168000, 129254400, 129340800, 129427200, 129513600, 129600000,
129686400, 129772800, 129859200, 129945600, 130032000, 130118400,
130204800, 130291200, 130377600, 130464000, 130550400, 130636800,
130723200, 130809600, 130896000, 130982400, 131068800, 131155200,
131241600, 131328000, 131414400, 131500800, 131587200, 131673600,
131760000, 131846400, 131932800, 132019200, 132105600, 132192000,
132278400, 132364800, 132451200, 132537600, 132624000, 132710400,
132796800, 132883200, 132969600, 133056000, 133142400, 133228800,
133315200, 133401600, 133488000, 133574400, 133660800, 133747200,
133833600, 133920000, 134006400, 134092800, 134179200, 134265600,
134352000, 134438400, 134524800, 134611200, 134697600, 134784000,
134870400, 134956800, 135043200, 135129600, 135216000, 135302400,
135388800, 135475200, 135561600, 135648000, 135734400, 135820800,
135907200, 135993600, 136080000, 136166400, 136252800, 136339200,
136425600, 136512000, 136598400, 136684800, 136771200, 136857600,
136944000, 137030400, 137116800, 137203200, 137289600, 137376000,
137462400, 137548800, 137635200, 137721600, 137808000, 137894400,
137980800, 138067200, 138153600, 138240000, 138326400, 138412800,
138499200, 138585600, 138672000, 138758400, 138844800, 138931200,
139017600, 139104000, 139190400, 139276800, 139363200, 139449600,
139536000, 139622400, 139708800, 139795200, 139881600, 139968000,
140054400, 140140800, 140227200, 140313600, 140400000, 140486400,
140572800, 140659200, 140745600, 140832000, 140918400, 141004800,
141091200, 141177600, 141264000, 141350400, 141436800, 141523200,
141609600, 141696000, 141782400, 141868800, 141955200, 142041600,
142128000, 142214400, 142300800, 142387200, 142473600, 142560000,
142646400, 142732800, 142819200, 142905600, 142992000, 143078400,
143164800, 143251200, 143337600, 143424000, 143510400, 143596800,
143683200, 143769600, 143856000, 143942400, 144028800, 144115200,
144201600, 144288000, 144374400, 144460800, 144547200, 144633600,
144720000, 144806400, 144892800, 144979200, 145065600, 145152000,
145238400, 145324800, 145411200, 145497600, 145584000, 145670400,
145756800, 145843200, 145929600, 146016000, 146102400, 146188800,
146275200, 146361600, 146448000, 146534400, 146620800, 146707200,
146793600, 146880000, 146966400, 147052800, 147139200, 147225600,
147312000, 147398400, 147484800, 147571200, 147657600, 147744000,
147830400, 147916800, 148003200, 148089600, 148176000, 148262400,
148348800, 148435200, 148521600, 148608000, 148694400, 148780800,
148867200, 148953600, 149040000, 149126400, 149212800, 149299200,
149385600, 149472000, 149558400, 149644800, 149731200, 149817600,
149904000, 149990400, 150076800, 150163200, 150249600, 150336000,
150422400, 150508800, 150595200, 150681600, 150768000, 150854400,
150940800, 151027200, 151113600, 151200000, 151286400, 151372800,
151459200, 151545600, 151632000, 151718400, 151804800, 151891200,
151977600, 152064000, 152150400, 152236800, 152323200, 152409600,
152496000, 152582400, 152668800, 152755200, 152841600, 152928000,
153014400, 153100800, 153187200, 153273600, 153360000, 153446400,
153532800, 153619200, 153705600, 153792000, 153878400, 153964800,
154051200, 154137600, 154224000, 154310400, 154396800, 154483200,
154569600, 154656000, 154742400, 154828800, 154915200, 155001600,
155088000, 155174400, 155260800, 155347200, 155433600, 155520000,
155606400, 155692800, 155779200, 155865600, 155952000, 156038400,
156124800, 156211200, 156297600, 156384000, 156470400, 156556800,
156643200, 156729600, 156816000, 156902400, 156988800, 157075200,
157161600, 157248000, 157334400, 157420800, 157507200, 157593600,
157680000), y = c(4.65, 4.65, 4.65, 4.48, 5.16, 5.52, 5.34, 5.34,
4.82, 4.65, 4.48, 4.31, 4.31, 4.31, 4.14, 3.82, 3.98, 3.98, 4.31,
5.71, 6.5, 6.3, 5.71, 5.71, 5.16, 4.65, 4.14, 3.98, 4.48, 4.48,
4.31, 4.65, 4.31, 3.98, 3.98, 3.98, 3.98, 3.98, 3.98, 3.82, 3.67,
3.67, 3.98, 3.98, 3.82, 3.82, 3.82, 4.14, 5.9, 4.48, 3.98, 3.98,
3.82, 3.67, 3.67, 3.67, 4.65, 3.98, 4.31, 4.31, 3.67, 4.31, 6.1,
7.3, 7.5, 7.5, 8.14, 10.8, 16.1, 14.8, 12.5, 9.9, 8.14, 6.9,
6.1, 5.34, 5.16, 4.99, 4.99, 4.99, 4.99, 5.52, 6.3, 7.3, 6.9,
5.9, 5.71, 5.71, 8.58, 31.5, 33.7, 18.4, 11.3, 16.1, 32.9, 45.3,
54, 25.7, 18, 15.9, 15.6, 14.5, 15.9, 35.9, 37.5, 29.4, 27.5,
30.1, 27.5, 30.8, 29.4, 22, 20.1, 35.9, 36.7, 32.9, 22, 18, 15.9,
15.2, 14.8, 13, 12.7, 12.5, 11, 9.68, 8.8, 7.92, 7.3, 6.9, 7.3,
10.3, 11, 11.3, 11.9, 12.5, 13.6, 12.2, 10.8, 9.9, 9.46, 8.8,
7.5, 7.1, 7.71, 7.1, 6.1, 5.34, 5.34, 5.34, 5.52, 5.52, 6.3,
6.7, 6.5, 5.9, 5.71, 5.9, 5.71, 5.52, 7.3, 7.5, 7.1, 7.3, 6.7,
6.9, 7.3, 7.5, 10.8, 11.6, 8.58, 7.92, 7.1, 6.7, 6.5, 6.1, 5.9,
5.9, 5.71, 5.52, 5.52, 5.52, 5.9, 5.9, 5.71, 5.52, 5.52, 5.34,
5.34, 5.52, 6.5, 6.5, 5.71, 5.34, 5.16, 4.99, 4.82, 4.82, 4.99,
4.82, 4.82, 4.82, 4.82, 4.82, 4.65, 4.48, 4.48, 4.31, 4.31, 4.14,
4.14, 4.31, 4.48, 4.31, 4.31, 4.31, 4.99, 5.71, 6.3, 6.1, 6.1,
5.9, 5.71, 5.52, 5.52, 5.52, 5.52, 5.52, 5.34, 5.34, 5.52, 5.52,
5.52, 5.34, 5.34, 5.52, 5.34, 5.52, 5.52, 5.34, 5.34, 5.34, 5.34,
5.71, 5.9, 6.3, 6.9, 7.5, 6.5, 6.1, 6.1, 5.9, 6.1, 6.1, 5.9,
6.5, 6.5, 6.1, 5.9, 5.9, 5.71, 5.9, 5.9, 5.71, 4.99, 4.65, 5.16,
5.34, 5.34, 4.65, 4.99, 5.71, 5.34, 5.34, 5.34, 5.34, 4.99, 5.34,
5.34, 5.34, 5.34, 5.52, 5.34, 5.52, 5.71, 6.5, 7.71, 6.9, 6.5,
6.7, 6.1, 5.9, 6.1, 5.9, 5.71, 7.92, 7.71, 7.1, 7.92, 5.34, 5.16,
8.14, 10.1, 7.92, 7.3, 6.9, 6.9, 6.9, 8.58, 7.3, 6.9, 7.3, 6.3,
5.16, 6.1, 5.52, 4.99, 5.34, 5.34, 5.34, 5.16, 5.71, 5.52, 5.52,
5.16, 4.82, 5.52, 6.1, 5.9, 5.71, 5.52, 5.16, 4.99, 4.48, 4.82,
5.16, 5.16, 5.16, 5.16, 5.16, 4.82, 4.65, 3.82, 4.14, 4.65, 4.65,
4.31, 4.31, 5.16, 5.16, 5.16, 5.16, 5.16, 4.99, 4.65, 5.16, 5.16,
5.16, 5.16, 5.16, 5.16, 5.16, 5.16, 5.34, 5.34)), .Names = c("x",
"y"), row.names = c(NA, -365L), class = c("data.table", "data.frame"
), .internal.selfref = <pointer: 0x0000000005860788>)
> sessionInfo()
R version 3.2.2 (2015-08-14)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 7 x64 (build 7601) Service Pack 1
locale:
[1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252 LC_MONETARY=English_United States.1252
[4] LC_NUMERIC=C LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] data.table_1.9.6 readxl_0.1.0 mgcv_1.8-7 nlme_3.1-121 scales_0.3.0 sos_1.3-8 brew_1.0-6 ggplot2_1.0.1
[9] MASS_7.3-43
loaded via a namespace (and not attached):
[1] Rcpp_0.12.1 lattice_0.20-33 digest_0.6.8 chron_2.3-47 grid_3.2.2 plyr_1.8.3 gtable_0.1.2 magrittr_1.5
[9] stringi_0.5-5 reshape2_1.4.1 Matrix_1.2-2 labeling_0.3 proto_0.3-10 tools_3.2.2 stringr_1.0.0 munsell_0.4.2
[17] colorspace_1.2-6
部分解决方案
我仍然不太清楚为什么这两种方法会给出不同的答案,这让我感到困扰。但是,在经过大量的互联网搜索后,我找到了以下解决方法:
library(readxl)
library(data.table)
library(ggplot2)
library(scales)
library(mgcv)
stackOF_data <- read_excel("C:/Users/jel4049/Desktop/mean-daily-flow-cumecs- vatnsdals.xlsx", sheet = "Data")
stackOF_data <- data.table(stackOF_data)
stackOF_data <- stackOF_data[,.(timeseries=as.POSIXct(Date,format("%Y-%m-%d")),mdf)]
a <- stackOF_data[,.(x=as.numeric(timeseries),y=mdf)]
a1 <- gam(y~s(x, k=100, bs="cs"),data=a)
a2=data.table(gam_mdf = predict(a1,a))
preds <- predict(a1,se.fit=TRUE)
my_data <- data.frame(mu=preds$fit, low =(preds$fit - 1.96 * preds$se.fit), high = (preds$fit + 1.96 * preds$se.fit))
m <- ggplot()+
geom_line(data = a2, aes(x=stackOF_data$timeseries, y=gam_mdf), size=1, col="blue")+
geom_smooth(data=my_data,aes(ymin = low, ymax = high, x=stackOF_data$timeseries, y = mu), stat = "identity", col="green")
m
现在至少我知道,我可以从一些mgcv函数中获取的摘要和数据拟合质量信息与我的图形匹配。
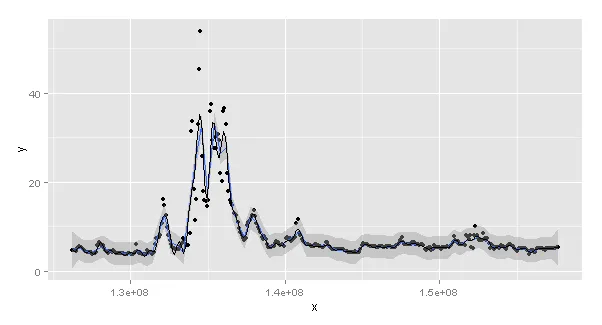
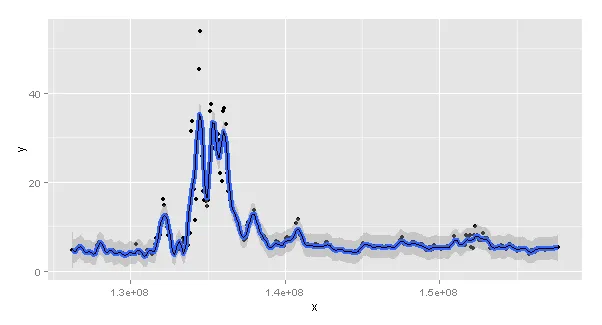
Error: Invalid input: time_trans works with objects of class POSIXct only。似乎在gam和stat_smooth对于可以接受的日期/时间信息的要求上存在差异。我不确定如何使它们相同,并且仍然拥有可读的x轴的图表...如果您有任何想法,我很乐意得到帮助!谢谢。 - CJ9w <- ggplot() + geom_line(data = a2, aes(x=a$x, y=gam_mdf), size=1, col="blue")+ geom_smooth(data = a, aes(x=x, y=y),method="gam", formula=y~s(x,k=100, bs="cs"), col="green",se=FALSE, size=1) w这应该更接近您的想法,对吧?但是图形仍然显示有差异。您有什么想法吗?谢谢。 - CJ9