如何根据给定的变量比例修改“点”的大小?
例如,使用ggplot可以这样做:
ggplot(mydata,aes(x=x, y=y))+geom_point(aes(size=mysize))+geom_line(aes(group=id, color=id))
我的size变量代表了我想要的每个点的大小。 但是当数据集很大时,这个过程非常缓慢。
我已经尝试使用lattice:
xyplot(y~x , type="b", col="black", col.line="blue", data=mydata, pch=16, lwd=1 )
但是我无法找到一种使用lattice的size=选项的有效方法。
我尝试了cex选项,但它产生了奇怪的结果。
这是我的数据的简化版本:(它是一个data.table,但如果您愿意,可以使用data.frame)
structure(list(id = c(3059L, 1161L, 3996L, 6330L, 6675L, 1511L,
3678L, 294L, 596L, 2440L, 446L, 2635L, 6073L, 3709L, 744L, 4997L,
3495L, 6447L, 6693L, 1040L, 1031L, 690L, 352L, 6311L, 2599L,
6425L, 3758L, 690L, 6742L, 3025L, 6348L, 214L, 222L, 8192L, 615L,
2939L, 5351L, 255L, 1531L, 6426L, 1686L, 2677L, 1919L, 3665L,
6514L, 630L, 820L, 2138L, 6695L, 1323L, 6246L, 2102L, 2600L,
3663L, 3851L, 970L, 1124L, 4071L, 1806L, 4579L, 3395L, 4371L,
1466L, 201L, 2112L, 8653L, 4407L, 1959L, 6341L, 2214L, 6515L,
1390L, 6346L, 5373L, 662L, 2198L, 1971L, 6177L, 4652L, 4420L,
6527L, 2704L, 6366L, 1111L, 6156L, 151L, 734L, 4286L, 5085L,
2359L, 2818L, 339L, 8486L, 5303L, 5076L, 8490L, 1230L, 1884L,
5204L, 2880L, 8463L, 215L, 6778L, 6329L, 5797L, 584L, 4831L,
4806L, 2581L, 3972L, 2298L, 3136L, 335L, 5538L, 1528L, 518L,
3552L, 3874L, 1967L, 4333L, 3035L, 4112L, 215L, 1768L, 866L,
3545L, 8085L, 8622L, 2844L, 2663L, 1356L, 4902L, 880L, 8219L,
1486L, 3086L, 685L, 417L, 5966L, 221L, 8401L, 8378L, 1542L, 1815L,
515L, 6262L, 3522L, 2440L, 182L, 946L, 3924L, 6219L, 2282L, 4165L,
4969L, 5829L, 6707L, 4467L, 8645L, 8267L, 3960L, 1432L, 8203L,
1691L, 5515L, 2005L, 5578L, 6732L, 103L, 740L, 8239L, 2037L,
630L, 8286L, 4005L, 3874L, 1691L, 201L, 4191L, 1259L, 4476L,
5210L, 798L, 5829L, 6499L, 120L, 1725L, 2185L, 3795L, 5918L,
3474L, 936L, 4625L, 4449L, 5579L, 5906L, 2050L, 3376L, 6140L,
8021L, 5009L, 3545L, 346L, 8513L, 1529L, 4155L, 3029L, 5785L,
397L, 1448L, 1804L, 6209L, 3043L, 2448L, 8371L, 5729L, 1602L,
5600L, 1488L, 280L, 1986L, 2858L, 1742L, 1939L, 4869L, 751L,
2967L, 6350L, 612L, 3935L, 3972L, 8321L, 8225L, 8117L, 5181L,
2214L, 1461L, 8150L, 3218L, 3699L, 4906L, 4488L, 1317L, 866L,
6832L, 8394L, 527L, 1384L, 3356L, 3320L, 1951L, 2979L, 3884L,
1664L, 8446L, 1851L, 1105L, 816L, 6323L, 4490L, 1020L, 3888L,
1086L, 4804L, 1203L, 3464L, 836L, 1591L, 755L, 6332L, 221L, 3519L,
1565L, 1169L, 2841L, 3992L, 5296L, 6187L, 3001L, 3711L, 1394L,
3333L, 1728L, 4320L, 6670L, 4653L, 6409L, 2369L, 8428L, 3714L,
8383L, 2142L, 1803L, 589L, 1986L, 866L, 866L, 4558L, 8139L, 3996L,
3667L, 2939L, 4626L, 3950L, 4371L, 6036L, 3212L, 8534L, 1599L,
1801L, 5015L, 121L, 8258L, 3811L, 1277L, 1161L, 4854L, 4581L,
8150L, 3223L, 2939L, 8548L, 1691L, 2969L, 1705L, 3477L, 5351L,
1120L, 3921L, 3915L, 2599L, 3457L, 630L, 2296L, 1569L, 6378L,
3447L, 6438L, 2697L, 2236L, 675L, 6147L, 4842L, 6205L, 4453L,
4478L, 2394L, 2115L, 5799L, 4683L, 1699L, 4757L, 8566L, 2125L,
3365L, 5194L, 1448L, 2043L, 2549L, 8647L, 1542L, 6179L, 1706L,
4099L, 5996L, 5289L, 6383L, 5102L, 488L, 889L, 3323L, 8461L,
1673L, 919L, 661L, 6505L, 1624L, 8261L, 6118L, 3935L, 2288L,
1230L, 1221L, 8359L, 8611L, 5710L, 6064L, 5238L, 2251L, 72L,
678L, 1649L, 8304L, 6056L, 8294L, 1967L, 5914L, 536L, 6330L,
8548L, 1111L, 3773L, 1381L, 3522L, 8461L, 1163L, 3111L, 3035L,
8101L, 1705L, 1690L, 2988L, 2243L, 3921L, 6348L, 4683L, 8424L,
3117L, 2015L, 559L, 8596L, 4235L, 280L, 8197L, 6096L, 3130L,
2880L, 1224L, 3636L, 8291L, 108L, 3213L, 2386L, 3350L, 3526L,
8116L, 2213L, 3519L, 1387L, 3672L, 4096L, 4288L, 905L, 3348L,
380L, 876L, 1685L, 8636L, 5559L, 1630L, 4696L, 5068L, 2351L,
757L, 3842L, 3517L, 8270L, 2880L, 8572L, 3384L, 6056L, 3884L,
1259L, 1053L, 6262L, 3965L, 4401L, 5295L, 4467L, 8216L, 8197L,
3500L, 5475L, 6662L, 2117L, 1092L, 5500L, 1447L, 6283L, 5930L,
2486L, 3258L, 4272L, 1616L, 3024L, 4104L, 3760L, 4431L, 5830L,
6366L, 5202L, 2868L, 5097L, 8647L, 6701L, 1167L, 5521L, 1209L,
5979L, 6178L), cars = structure(c(11L, 12L, 12L, 11L, 12L, 11L,
12L, 9L, 1L, 9L, 8L, 13L, 12L, 12L, 12L, 9L, 12L, 10L, 8L, 10L,
9L, 12L, 13L, 8L, 10L, 9L, 7L, 9L, 11L, 13L, 9L, 3L, 12L, 13L,
13L, 12L, 11L, 13L, 13L, 8L, 7L, 13L, 13L, 13L, 13L, 12L, 7L,
9L, 11L, 10L, 11L, 9L, 12L, 12L, 12L, 13L, 13L, 11L, 6L, 12L,
13L, 9L, 9L, 2L, 11L, 11L, 13L, 12L, 13L, 8L, 11L, 9L, 11L, 11L,
7L, 12L, 10L, 7L, 13L, 5L, 12L, 10L, 7L, 13L, 13L, 12L, 7L, 9L,
10L, 12L, 13L, 13L, 7L, 13L, 12L, 13L, 13L, 9L, 13L, 9L, 10L,
10L, 13L, 11L, 13L, 13L, 13L, 13L, 13L, 11L, 11L, 7L, 6L, 13L,
11L, 7L, 12L, 10L, 4L, 11L, 13L, 12L, 6L, 8L, 12L, 7L, 12L, 12L,
12L, 10L, 8L, 9L, 11L, 13L, 12L, 9L, 13L, 10L, 12L, 8L, 7L, 12L,
13L, 13L, 13L, 13L, 13L, 10L, 12L, 3L, 13L, 13L, 12L, 13L, 13L,
9L, 6L, 11L, 11L, 12L, 13L, 12L, 13L, 6L, 8L, 13L, 10L, 13L,
11L, 13L, 11L, 7L, 12L, 13L, 13L, 11L, 4L, 11L, 11L, 5L, 11L,
12L, 13L, 11L, 8L, 9L, 10L, 11L, 12L, 3L, 10L, 8L, 10L, 13L,
8L, 13L, 6L, 12L, 13L, 12L, 13L, 6L, 12L, 12L, 11L, 13L, 10L,
12L, 10L, 12L, 8L, 10L, 10L, 13L, 13L, 13L, 8L, 13L, 8L, 12L,
7L, 8L, 11L, 1L, 11L, 12L, 7L, 12L, 13L, 11L, 7L, 13L, 12L, 9L,
13L, 13L, 2L, 4L, 13L, 12L, 9L, 10L, 11L, 12L, 9L, 13L, 10L,
12L, 9L, 7L, 12L, 13L, 11L, 13L, 7L, 4L, 7L, 13L, 13L, 13L, 13L,
12L, 8L, 5L, 8L, 13L, 6L, 13L, 12L, 8L, 12L, 6L, 8L, 6L, 13L,
12L, 8L, 6L, 11L, 8L, 4L, 12L, 10L, 12L, 12L, 11L, 8L, 12L, 10L,
9L, 13L, 12L, 10L, 11L, 13L, 12L, 12L, 12L, 11L, 7L, 13L, 12L,
11L, 7L, 8L, 13L, 12L, 13L, 12L, 13L, 13L, 13L, 13L, 11L, 13L,
13L, 6L, 5L, 9L, 13L, 10L, 8L, 13L, 8L, 10L, 12L, 10L, 11L, 11L,
13L, 13L, 8L, 13L, 13L, 12L, 2L, 12L, 1L, 5L, 13L, 4L, 12L, 3L,
3L, 13L, 8L, 13L, 13L, 10L, 10L, 5L, 11L, 11L, 12L, 10L, 8L,
13L, 5L, 13L, 11L, 7L, 12L, 9L, 13L, 12L, 5L, 12L, 12L, 7L, 10L,
3L, 12L, 11L, 6L, 11L, 12L, 12L, 12L, 11L, 7L, 8L, 11L, 10L,
11L, 13L, 13L, 13L, 12L, 6L, 12L, 11L, 13L, 9L, 8L, 13L, 7L,
4L, 13L, 9L, 9L, 10L, 12L, 1L, 12L, 12L, 13L, 9L, 12L, 7L, 6L,
13L, 9L, 13L, 10L, 12L, 12L, 13L, 13L, 12L, 13L, 12L, 7L, 13L,
3L, 12L, 12L, 13L, 2L, 5L, 9L, 12L, 10L, 7L, 12L, 12L, 11L, 10L,
13L, 13L, 13L, 11L, 13L, 13L, 12L, 10L, 7L, 13L, 12L, 6L, 13L,
8L, 11L, 8L, 13L, 10L, 13L, 13L, 11L, 8L, 11L, 8L, 7L, 11L, 5L,
11L, 13L, 11L, 13L, 13L, 12L, 6L, 13L, 9L, 13L, 8L, 6L, 8L, 6L,
13L, 9L, 13L, 12L, 6L, 8L, 12L, 10L, 13L, 12L, 12L, 11L, 8L,
11L, 11L, 2L, 11L, 12L, 11L, 11L, 6L, 12L), .Label = c("FORD",
"VW", "PEUGEOT", "RENAULT", "TOYOTA", "BMW", "NISSAN", "MB",
"AUDI", "HONDA", "FIAT", "LR", "SKODA", "MAZDA", "MINI", "KIA",
"VOLVO", "SEAT", "SUZUKI", "MITSU", "JAGUAR", "ROVER", "SAAB",
"LEXUS", "CHEVRO", "MG", "PORSCHE"), class = "factor"), numb = c(1L,
1L, 1L, 1L, 1L, 2L, 1L, 3L, 2L, 2L, 2L, 1L, 1L, 1L, 5L, 2L, 2L,
1L, 2L, 3L, 1L, 4L, 3L, 3L, 1L, 1L, 1L, 2L, 2L, 1L, 5L, 1L, 1L,
4L, 6L, 1L, 2L, 1L, 1L, 1L, 2L, 2L, 2L, 1L, 4L, 12L, 9L, 1L,
1L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 1L, 1L, 2L, 1L, 3L, 2L, 1L, 6L,
1L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 2L, 1L, 4L, 2L, 1L, 1L, 1L, 1L,
2L, 2L, 2L, 7L, 1L, 2L, 6L, 1L, 1L, 1L, 3L, 2L, 2L, 6L, 1L, 1L,
2L, 2L, 3L, 2L, 2L, 1L, 2L, 2L, 3L, 3L, 4L, 1L, 2L, 2L, 1L, 5L,
3L, 1L, 1L, 1L, 1L, 2L, 2L, 1L, 3L, 1L, 2L, 1L, 10L, 4L, 1L,
2L, 3L, 1L, 2L, 3L, 1L, 1L, 1L, 2L, 8L, 1L, 1L, 4L, 2L, 1L, 5L,
1L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 1L, 2L, 1L, 3L, 1L,
1L, 2L, 1L, 2L, 3L, 4L, 2L, 1L, 2L, 1L, 3L, 1L, 2L, 8L, 1L, 1L,
3L, 6L, 4L, 1L, 2L, 1L, 1L, 3L, 1L, 2L, 1L, 1L, 1L, 1L, 4L, 1L,
1L, 2L, 2L, 2L, 1L, 3L, 1L, 2L, 2L, 1L, 5L, 1L, 1L, 1L, 2L, 6L,
2L, 1L, 4L, 1L, 3L, 2L, 1L, 3L, 2L, 1L, 3L, 2L, 2L, 2L, 2L, 1L,
3L, 1L, 1L, 2L, 2L, 1L, 2L, 1L, 3L, 1L, 2L, 2L, 3L, 3L, 3L, 1L,
4L, 1L, 2L, 2L, 7L, 2L, 1L, 1L, 3L, 1L, 2L, 1L, 1L, 1L, 1L, 1L,
3L, 6L, 3L, 2L, 8L, 3L, 2L, 1L, 3L, 1L, 1L, 1L, 2L, 1L, 2L, 2L,
1L, 4L, 2L, 2L, 3L, 3L, 2L, 1L, 2L, 2L, 1L, 3L, 1L, 1L, 1L, 3L,
2L, 3L, 1L, 2L, 1L, 1L, 1L, 3L, 1L, 4L, 1L, 2L, 2L, 2L, 2L, 1L,
2L, 3L, 4L, 2L, 1L, 1L, 1L, 2L, 1L, 4L, 1L, 3L, 2L, 1L, 4L, 5L,
5L, 3L, 2L, 8L, 1L, 3L, 4L, 1L, 2L, 1L, 2L, 2L, 4L, 13L, 1L,
4L, 2L, 1L, 1L, 1L, 4L, 1L, 1L, 1L, 2L, 1L, 2L, 3L, 1L, 1L, 5L,
5L, 1L, 2L, 1L, 1L, 1L, 6L, 3L, 3L, 1L, 4L, 1L, 3L, 2L, 1L, 2L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 5L, 2L, 1L, 3L, 2L, 2L, 3L, 2L, 4L, 4L, 2L, 1L, 2L, 2L,
2L, 1L, 2L, 1L, 7L, 1L, 2L, 2L, 1L, 2L, 1L, 2L, 1L, 1L, 4L, 2L,
16L, 4L, 2L, 6L, 1L, 2L, 3L, 4L, 3L, 5L, 1L, 1L, 6L, 3L, 5L,
3L, 1L, 1L, 3L, 1L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 3L, 3L, 1L, 1L,
2L, 1L, 1L, 2L, 3L, 2L, 1L, 1L, 2L, 1L, 2L, 2L, 1L, 1L, 2L, 1L,
2L, 1L, 6L, 1L, 1L, 2L, 1L, 5L, 1L, 6L, 4L, 1L, 1L, 2L, 3L, 1L,
2L, 1L, 1L, 2L, 1L, 6L, 1L, 3L, 4L, 1L, 2L, 3L, 1L, 5L, 2L, 1L,
2L, 2L, 2L, 1L, 1L, 1L, 1L), mysize = c(0.196195449459157, 0.259604625139873,
0.259604625139873, 0.196195449459157, 0.259604625139873, 0.0982578397212544,
0.259604625139873, 0.0761772853185596, 0.00627177700348432, 0.0759581881533101,
0.0655052264808362, 0.198433420365535, 0.259604625139873, 0.259604625139873,
0.167701863354037, 0.0759581881533101, 0.212543554006969, 0.120477433793361,
0.0655052264808362, 0.0664819944598338, 0.061917195076464, 0.183673469387755,
0.307479224376731, 0.0512465373961219, 0.120477433793361, 0.061917195076464,
0.0484893696381947, 0.0759581881533101, 0.0982578397212544, 0.198433420365535,
0.142857142857143, 0.0115628496829541, 0.259604625139873, 0.34402332361516,
0.375, 0.259604625139873, 0.0982578397212544, 0.198433420365535,
0.198433420365535, 0.0481163744871317, 0.0522648083623693, 0.282229965156794,
0.282229965156794, 0.198433420365535, 0.34402332361516, 0.5,
0.1, 0.061917195076464, 0.196195449459157, 0.120477433793361,
0.196195449459157, 0.0759581881533101, 0.212543554006969, 0.259604625139873,
0.212543554006969, 0.198433420365535, 0.198433420365535, 0.196195449459157,
0.0662020905923345, 0.259604625139873, 0.307479224376731, 0.0759581881533101,
0.061917195076464, 0.0625, 0.196195449459157, 0.0982578397212544,
0.282229965156794, 0.259604625139873, 0.282229965156794, 0.0481163744871317,
0.0982578397212544, 0.061917195076464, 0.0982578397212544, 0.196195449459157,
0.0699708454810496, 0.212543554006969, 0.120477433793361, 0.0484893696381947,
0.198433420365535, 0.00857888847444983, 0.212543554006969, 0.0682926829268293,
0.0522648083623693, 0.444444444444444, 0.198433420365535, 0.212543554006969,
0.0875, 0.061917195076464, 0.120477433793361, 0.259604625139873,
0.307479224376731, 0.282229965156794, 0.0522648083623693, 0.375,
0.259604625139873, 0.198433420365535, 0.282229965156794, 0.0759581881533101,
0.307479224376731, 0.0759581881533101, 0.0682926829268293, 0.120477433793361,
0.282229965156794, 0.0982578397212544, 0.307479224376731, 0.307479224376731,
0.34402332361516, 0.198433420365535, 0.282229965156794, 0.0982578397212544,
0.196195449459157, 0.0496894409937888, 0.054016620498615, 0.198433420365535,
0.196195449459157, 0.0484893696381947, 0.259604625139873, 0.0682926829268293,
0.0229965156794425, 0.196195449459157, 0.307479224376731, 0.259604625139873,
0.0662020905923345, 0.0481163744871317, 0.4, 0.0699708454810496,
0.259604625139873, 0.212543554006969, 0.21606648199446, 0.120477433793361,
0.0655052264808362, 0.0761772853185596, 0.196195449459157, 0.198433420365535,
0.259604625139873, 0.0759581881533101, 0.5, 0.120477433793361,
0.259604625139873, 0.0728862973760933, 0.0522648083623693, 0.259604625139873,
0.322981366459627, 0.198433420365535, 0.282229965156794, 0.282229965156794,
0.282229965156794, 0.120477433793361, 0.259604625139873, 0.0115628496829541,
0.198433420365535, 0.307479224376731, 0.21606648199446, 0.307479224376731,
0.198433420365535, 0.0759581881533101, 0.0354345393509884, 0.0775623268698061,
0.196195449459157, 0.259604625139873, 0.282229965156794, 0.259604625139873,
0.282229965156794, 0.054016620498615, 0.0728862973760933, 0.282229965156794,
0.120477433793361, 0.282229965156794, 0.196195449459157, 0.307479224376731,
0.196195449459157, 0.0522648083623693, 0.1, 0.198433420365535,
0.198433420365535, 0.0775623268698061, 0.0125, 0.032069970845481,
0.196195449459157, 0.0229965156794425, 0.196195449459157, 0.259604625139873,
0.307479224376731, 0.196195449459157, 0.0655052264808362, 0.061917195076464,
0.120477433793361, 0.196195449459157, 0.259604625139873, 0.0204081632653061,
0.120477433793361, 0.0481163744871317, 0.0682926829268293, 0.282229965156794,
0.0655052264808362, 0.198433420365535, 0.054016620498615, 0.259604625139873,
0.282229965156794, 0.212543554006969, 0.198433420365535, 0.0683229813664596,
0.259604625139873, 0.259604625139873, 0.196195449459157, 0.282229965156794,
0.0375, 0.212543554006969, 0.120477433793361, 0.183673469387755,
0.0481163744871317, 0.0664819944598338, 0.0682926829268293, 0.198433420365535,
0.307479224376731, 0.282229965156794, 0.0481163744871317, 0.307479224376731,
0.0655052264808362, 0.212543554006969, 0.0522648083623693, 0.0655052264808362,
0.196195449459157, 0.0138504155124654, 0.196195449459157, 0.259604625139873,
0.0522648083623693, 0.212543554006969, 0.198433420365535, 0.0982578397212544,
0.0484893696381947, 0.307479224376731, 0.259604625139873, 0.0759581881533101,
0.282229965156794, 0.307479224376731, 0.0152354570637119, 0.0193905817174515,
0.198433420365535, 0.183673469387755, 0.061917195076464, 0.0682926829268293,
0.0982578397212544, 0.2, 0.0759581881533101, 0.198433420365535,
0.120477433793361, 0.21606648199446, 0.061917195076464, 0.0522648083623693,
0.259604625139873, 0.198433420365535, 0.196195449459157, 0.198433420365535,
0.0484893696381947, 0.0193905817174515, 0.0875, 0.307479224376731,
0.282229965156794, 0.5, 0.307479224376731, 0.212543554006969,
0.0481163744871317, 0.0318559556786704, 0.0481163744871317, 0.198433420365535,
0.0354345393509884, 0.282229965156794, 0.259604625139873, 0.0655052264808362,
0.212543554006969, 0.0354345393509884, 0.0728862973760933, 0.0662020905923345,
0.282229965156794, 0.21606648199446, 0.0512465373961219, 0.0662020905923345,
0.196195449459157, 0.0655052264808362, 0.0229965156794425, 0.259604625139873,
0.0664819944598338, 0.259604625139873, 0.259604625139873, 0.196195449459157,
0.0512465373961219, 0.212543554006969, 0.0664819944598338, 0.061917195076464,
0.282229965156794, 0.259604625139873, 0.120477433793361, 0.196195449459157,
0.307479224376731, 0.259604625139873, 0.183673469387755, 0.259604625139873,
0.0982578397212544, 0.0522648083623693, 0.282229965156794, 0.212543554006969,
0.196195449459157, 0.0522648083623693, 0.0512465373961219, 0.34402332361516,
0.212543554006969, 0.198433420365535, 0.259604625139873, 0.198433420365535,
0.282229965156794, 0.198433420365535, 0.34402332361516, 0.196195449459157,
0.307479224376731, 0.282229965156794, 0.0354345393509884, 0.0291545189504373,
0.142857142857143, 0.322981366459627, 0.0664819944598338, 0.0655052264808362,
0.5, 0.0481163744871317, 0.0664819944598338, 0.183673469387755,
0.120477433793361, 0.0982578397212544, 0.196195449459157, 0.282229965156794,
0.282229965156794, 0.0728862973760933, 0.5, 0.198433420365535,
0.183673469387755, 0.0132404181184669, 0.259604625139873, 0.00484893696381947,
0.00857888847444983, 0.34402332361516, 0.00484893696381947, 0.259604625139873,
0.0115628496829541, 0.0132404181184669, 0.198433420365535, 0.0655052264808362,
0.307479224376731, 0.198433420365535, 0.120477433793361, 0.031055900621118,
0.0248447204968944, 0.196195449459157, 0.0982578397212544, 0.259604625139873,
0.120477433793361, 0.0481163744871317, 0.375, 0.0318559556786704,
0.307479224376731, 0.196195449459157, 0.0699708454810496, 0.259604625139873,
0.0761772853185596, 0.282229965156794, 0.259604625139873, 0.0229965156794425,
0.259604625139873, 0.259604625139873, 0.0484893696381947, 0.120477433793361,
0.0115628496829541, 0.259604625139873, 0.196195449459157, 0.0354345393509884,
0.196195449459157, 0.212543554006969, 0.259604625139873, 0.259604625139873,
0.196195449459157, 0.0484893696381947, 0.0481163744871317, 0.196195449459157,
0.120477433793361, 0.196195449459157, 0.322981366459627, 0.282229965156794,
0.198433420365535, 0.21606648199446, 0.0662020905923345, 0.212543554006969,
0.0775623268698061, 0.282229965156794, 0.0787172011661808, 0.0728862973760933,
0.282229965156794, 0.0484893696381947, 0.0229965156794425, 0.282229965156794,
0.0759581881533101, 0.061917195076464, 0.0682926829268293, 0.259604625139873,
0.0222222222222222, 0.259604625139873, 0.212543554006969, 0.282229965156794,
0.061917195076464, 0.212543554006969, 0.0484893696381947, 0.0662020905923345,
0.198433420365535, 0.061917195076464, 0.34402332361516, 0.0682926829268293,
1, 0.183673469387755, 0.282229965156794, 0.375, 0.259604625139873,
0.282229965156794, 0.21606648199446, 0.0699708454810496, 0.307479224376731,
0.0124223602484472, 0.259604625139873, 0.259604625139873, 0.375,
0.0152354570637119, 0.0248447204968944, 0.0761772853185596, 0.259604625139873,
0.120477433793361, 0.0595567867036011, 0.259604625139873, 0.259604625139873,
0.196195449459157, 0.120477433793361, 0.282229965156794, 0.198433420365535,
0.198433420365535, 0.0982578397212544, 0.307479224376731, 0.307479224376731,
0.259604625139873, 0.120477433793361, 0.0522648083623693, 0.198433420365535,
0.259604625139873, 0.0662020905923345, 0.307479224376731, 0.0655052264808362,
0.196195449459157, 0.0481163744871317, 0.282229965156794, 0.120477433793361,
0.282229965156794, 0.282229965156794, 0.196195449459157, 0.0481163744871317,
0.0982578397212544, 0.0481163744871317, 0.0522648083623693, 0.196195449459157,
0.05, 0.196195449459157, 0.198433420365535, 0.0982578397212544,
0.198433420365535, 0.322981366459627, 0.259604625139873, 0.05,
0.34402332361516, 0.061917195076464, 0.198433420365535, 0.0655052264808362,
0.054016620498615, 0.0481163744871317, 0.0662020905923345, 0.198433420365535,
0.061917195076464, 0.282229965156794, 0.259604625139873, 0.05,
0.0481163744871317, 0.21606648199446, 0.043731778425656, 0.198433420365535,
0.212543554006969, 0.21606648199446, 0.196195449459157, 0.062111801242236,
0.0982578397212544, 0.196195449459157, 0.0132404181184669, 0.0982578397212544,
0.212543554006969, 0.196195449459157, 0.196195449459157, 0.0354345393509884,
0.259604625139873)), .Names = c("id", "cars", "numb", "mysize"
), class = c("data.table", "data.frame"), row.names = c(NA, -500L
), .internal.selfref = <pointer: 0x0000000000120788>)
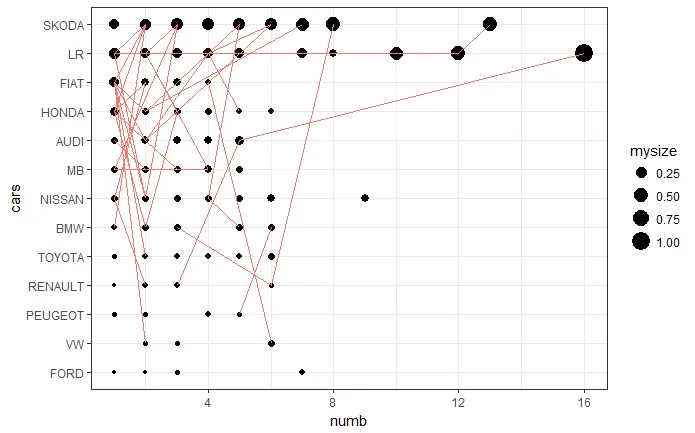
以下是使用ggplot绘制的图表
ggplot(mydata,aes(x=numb, y=cars))+geom_point(aes(size=mysize))+geom_line(aes(group=id, color="blue"), show.legend = FALSE)+theme_bw()
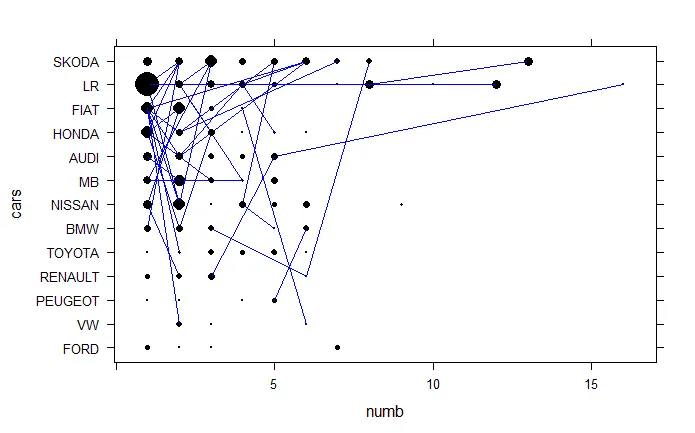
以及使用Lattice的绘图
xyplot(cars~numb , type="b", col="black", col.line="blue", data=mydata, pch=16, cex= mydata$mysize*3, lwd=1 , groups= mydata$id)
从上面的图片可以看出这些圆并不相等。 我不知道哪一个是错误的。
PD2: 我已经汇总了数据,只保留了唯一的对。
poi <- unique(mydata, by=c("cars","numb"))
structure(list(id = c(3059L, 1161L, 1511L, 294L, 596L, 2440L,
446L, 2635L, 744L, 3495L, 6447L, 1040L, 1031L, 690L, 352L, 6311L,
3758L, 6348L, 214L, 8192L, 615L, 6426L, 1686L, 2677L, 630L, 820L,
1806L, 201L, 662L, 4420L, 2704L, 1111L, 734L, 3136L, 335L, 1967L,
866L, 2844L, 685L, 221L, 1542L, 6707L, 4467L, 630L, 1691L, 201L,
1259L, 5918L, 3545L, 3029L, 1939L, 1461L, 8150L, 866L, 4804L,
4581L, 630L, 6378L, 6438L, 675L, 6205L, 4683L, 1699L, 8304L,
1381L, 6348L, 8197L, 2386L, 1053L, 8197L, 4104L, 5202L), cars = structure(c(11L,
12L, 11L, 9L, 1L, 9L, 8L, 13L, 12L, 12L, 10L, 10L, 9L, 12L, 13L,
8L, 7L, 9L, 3L, 13L, 13L, 8L, 7L, 13L, 12L, 7L, 6L, 2L, 7L, 5L,
10L, 13L, 7L, 7L, 6L, 4L, 12L, 12L, 13L, 8L, 13L, 6L, 11L, 12L,
4L, 11L, 5L, 3L, 6L, 10L, 1L, 2L, 4L, 12L, 5L, 5L, 13L, 2L, 1L,
4L, 3L, 10L, 5L, 9L, 1L, 12L, 3L, 7L, 5L, 6L, 10L, 8L), .Label = c("FORD",
"VW", "PEUGEOT", "RENAULT", "TOYOTA", "BMW", "NISSAN", "MB",
"AUDI", "HONDA", "FIAT", "LR", "SKODA", "MAZDA", "MINI", "KIA",
"VOLVO", "SEAT", "SUZUKI", "MITSU", "JAGUAR", "ROVER", "SAAB",
"LEXUS", "CHEVRO", "MG", "PORSCHE"), class = "factor"), numb = c(1L,
1L, 2L, 3L, 2L, 2L, 2L, 1L, 5L, 2L, 1L, 3L, 1L, 4L, 3L, 3L, 1L,
5L, 1L, 4L, 6L, 1L, 2L, 2L, 12L, 9L, 2L, 6L, 4L, 1L, 2L, 7L,
6L, 5L, 3L, 2L, 10L, 3L, 8L, 4L, 5L, 1L, 3L, 8L, 6L, 4L, 2L,
4L, 5L, 6L, 3L, 3L, 3L, 7L, 3L, 4L, 13L, 2L, 1L, 1L, 2L, 5L,
5L, 4L, 7L, 16L, 5L, 3L, 6L, 6L, 4L, 5L), mysize = c(0.196195449459157,
0.259604625139873, 0.0982578397212544, 0.0761772853185596, 0.00627177700348432,
0.0759581881533101, 0.0655052264808362, 0.198433420365535, 0.167701863354037,
0.212543554006969, 0.120477433793361, 0.0664819944598338, 0.061917195076464,
0.183673469387755, 0.307479224376731, 0.0512465373961219, 0.0484893696381947,
0.142857142857143, 0.0115628496829541, 0.34402332361516, 0.375,
0.0481163744871317, 0.0522648083623693, 0.282229965156794, 0.5,
0.1, 0.0662020905923345, 0.0625, 0.0699708454810496, 0.00857888847444983,
0.0682926829268293, 0.444444444444444, 0.0875, 0.0496894409937888,
0.054016620498615, 0.0229965156794425, 0.4, 0.21606648199446,
0.5, 0.0728862973760933, 0.322981366459627, 0.0354345393509884,
0.0775623268698061, 0.1, 0.0125, 0.032069970845481, 0.0229965156794425,
0.0204081632653061, 0.0683229813664596, 0.0375, 0.0138504155124654,
0.0152354570637119, 0.0193905817174515, 0.2, 0.0318559556786704,
0.0291545189504373, 0.5, 0.0132404181184669, 0.00484893696381947,
0.00484893696381947, 0.0132404181184669, 0.031055900621118, 0.0248447204968944,
0.0787172011661808, 0.0222222222222222, 1, 0.0124223602484472,
0.0595567867036011, 0.05, 0.05, 0.043731778425656, 0.062111801242236
)), class = c("data.table", "data.frame"), row.names = c(NA,
-72L), .Names = c("id", "cars", "numb", "mysize"))
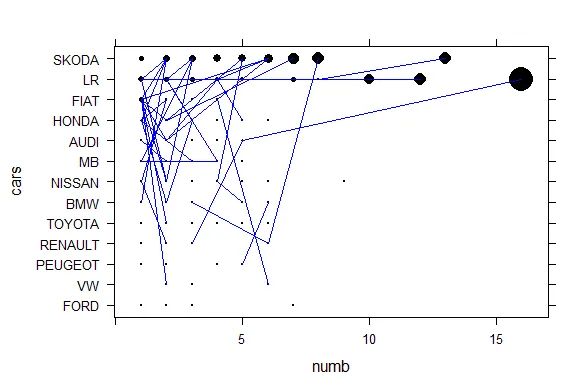
Lattice 对于这个去重后的数据集并没有产生完全相同的结果,但至少接近我们想要的结果。
p1 <- xyplot(cars~numb , type="p", col="black", data=poi, pch=16, cex= poi$mysize*3)
p2 <- xyplot(cars ~ numb , type = "l", col.line = "blue", data = mydata, lwd = 1 , groups = mydata$id)
p1+as.layer(p2)
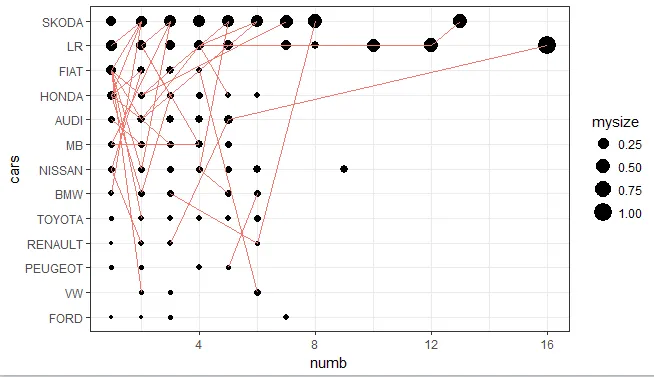
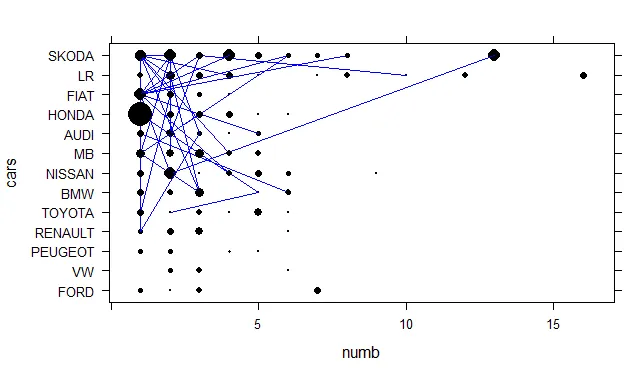
library(dplyr) ; nrow(mydata %>% filter(cars=="LR" & numb<2) %>% droplevels()),您将获得61行。我想知道这是否在起作用。 - KoenVlattice绘图中起到一定作用,但在ggplot中没有起作用。我将使用代码和图形更新我的答案。您应该考虑这是否是您想要的:多次(每个点不同)重叠一个点并且看不到它(ggplot),或者看到点的大小增加(lattice)。也许您需要一种不同的可视化方式。 - KoenV