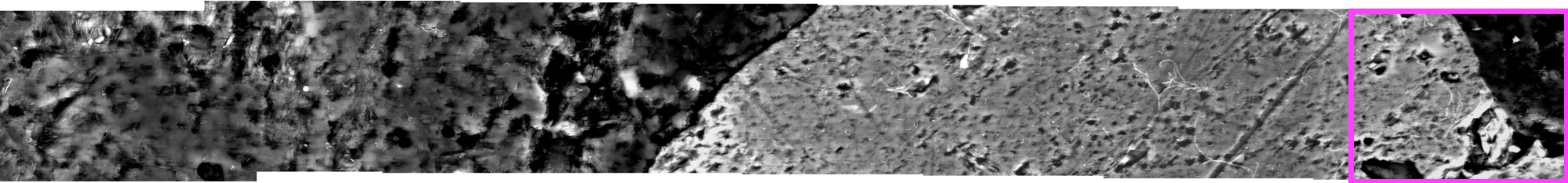
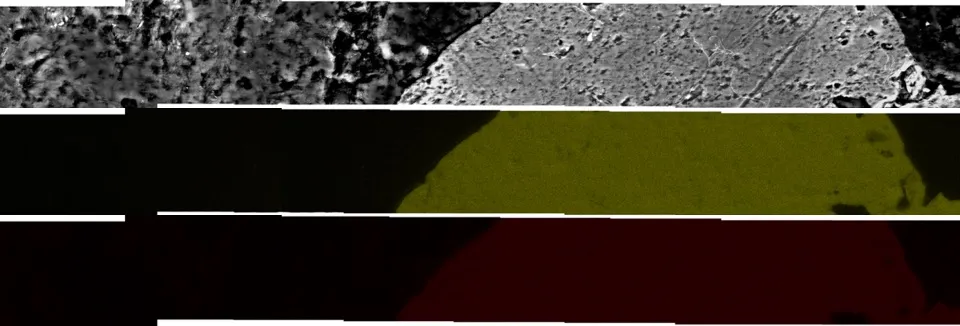
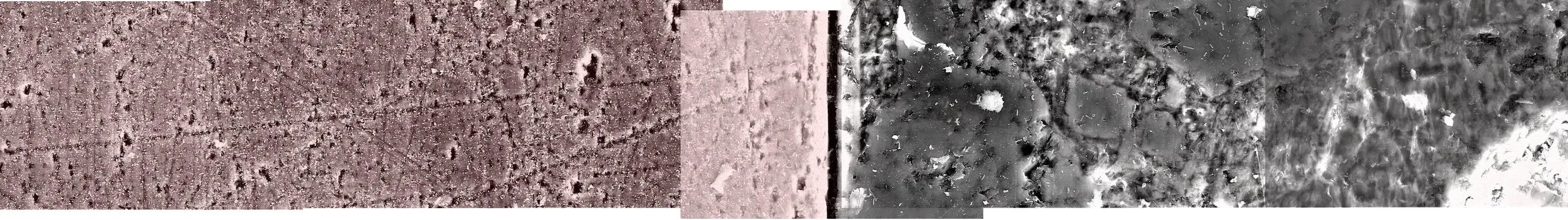
很希望能够完全使用Photoshop脚本或完全使用R来完成这个任务,但我有一个使用两者的工作解决方案。我手动从Photoshop的photomerge输出中提取了每个图层左下角的XY坐标,并将它们分配给了R中的光栅图像。
message("
##### ######
##### ## ##### ## # # ##### # ##### #### # # # #
# # # # # # # # # # # # # # # # #
# # # # # # # ##### # # # # ###### ######
# # ###### # ###### # # # # # # # # #
# # # # # # # # # # # # # # # # # #
##### # # # # # ##### # # # #### # # # #
")
message("DataStitchR was created by Caitlin Casar and is maintained at github.com/CaitlinCasar/dataStitchR.
")
message("DataStitchR stitches panoramic images of SEM images coupled to x-ray energy dispersive spectroscopy.
")
message("For help, run 'dataStitchR.R --help'.
")
suppressPackageStartupMessages(require(optparse))
option_list = list(
make_option(c("-f", "--file"), action="store", default=getwd(), type='character',
help="Input parent directory with subdirectories of element xray data to be stitched. The element names should be abbreviated, i.e. 'Ca' for calcium."),
make_option(c("-o", "--out"), action="store", default=NA, type='character',
help="Output file directory. This is where your x-ray raster brick and output figures will be saved."),
make_option(c("-n", "--name"), action="store", default=NA, type='character',
help="Optional name for output files."),
make_option(c("-b", "--base-images"), action="store", default=NA, type='character',
help="SEM image file directory."),
make_option(c("-c", "--coords"), action="store", default=NA, type='character',
help="Tab-delimited file of xy coordinates for each image. A third column should denote stitching positions that correspond to the file names for each image."),
make_option(c("-u", "--use-positions"), action="store", default="-?(?<![Kα1||Kα1_2])\\d+", type='character',
help="Optional regex pattern to extract position IDs from each file name that corresponds to positions in the xy file. The default searches for numbers that appear after 'Kα1' or 'Kα2'. Numbers can include signs, i.e. -1 is acceptable."),
make_option(c("-z", "--z-format"), action="store", default="*", type='character',
help="Optional regex pattern of x-ray image formats to select for stitching, i.e. '.tif'."),
make_option(c("-m", "--make"), action="store", default="*", type='character',
help="Optional regex pattern of SEM image formats to select for stitching, i.e. '.tif'. You do not need to specify this unless you are generating a PDF output."),
make_option(c("-a", "--all-exclude"), action="store", default=NA, type='character',
help="Optional regex pattern of x-ray file directories to exclude from stitiching, i.e. the element your sample was coated with."),
make_option(c("-d", "--drop"), action="store", default=NA, type='character',
help="Optional regex pattern of files to exclude from x-ray data stitching."),
make_option(c("-y", "--y-exclude"), action="store", default=NA, type='character',
help="Optional regex pattern of files to exclude from SEM image stitiching. You do not need to specify this unless you are generating a PDF output."),
make_option(c("-v", "--verbose"), action="store_true", default=TRUE,
help="Print updates to console [default %default]."),
make_option(c("-q", "--quiet"), action="store_false", dest="verbose",
help="Do not print anything to the console."),
make_option(c("-p", "--pdf"), action="store", default=FALSE,
help="Generate PDF of x-ray brick colored by element superimposed on the SEM image, default is TRUE [default %default].")
)
opt = parse_args(OptionParser(option_list=option_list))
suppressPackageStartupMessages(require(pacman))
pacman::p_load(raster, magick, tidyverse, rasterVis, ggnewscale, Hmisc, cowplot)
directories <- list.dirs(opt$f, full.names = T , recursive =F)
if(!is.na(opt$a)){
directories <- directories[!str_detect(directories, opt$a)]
}
xy <- read_delim(opt$c, delim = "\t", col_types = cols())
positions <- xy %>%
select(-x, -y)
xray_brick_list <- list()
xray_data <- list()
for(j in 1:length(directories)){
path = directories[j]
files <- list.files(path, full.names = T, pattern = opt$z)
if(!is.na(opt$d)){
files <- files[!str_detect(files, opt$d)]
}
if(length(files) >0){
xray_data[[j]] <- str_extract(path, "([^/]+$)")
message(paste0("Stitching ", str_extract(xray_data[[j]], "([^/]+$)"), " data (element ", j, " of ", length(directories), ")..."))
xy_id <- which(positions[[1]] %in% str_extract(files, opt$u))
panorama <- list()
for(i in 1:length(files)){
message(paste0("Processing image ", i, " of ", length(files), "..."))
image <- files[i] %>% image_read() %>%
image_quantize(colorspace = 'gray') %>%
image_equalize()
temp_file <- tempfile()
image_write(image, path = temp_file, format = 'tiff')
image <- raster(temp_file) %>%
cut(breaks = c(-Inf, 150, Inf)) - 1
image <- aggregate(image, fact=4)
image_extent <- extent(matrix(c(xy$x[xy_id[i]], xy$x[xy_id[i]] + 1024, xy$y[xy_id[i]], xy$y[xy_id[i]]+704), nrow = 2, ncol = 2, byrow = T))
image_raster <- setExtent(raster(nrows = 704, ncols = 1024), image_extent, keepres = F)
values(image_raster) <- values(image)
panorama[[xy_id[i]]] <- image_raster
}
empty_xy_id <- which(!positions[[1]] %in% str_extract(files, opt$u))
if(length(empty_xy_id) > 0){
for(k in 1:length(empty_xy_id)){
empty_raster_extent <- extent(matrix(c(xy$x[empty_xy_id[k]], xy$x[empty_xy_id[k]] + 1024, xy$y[empty_xy_id[k]], xy$y[empty_xy_id[k]]+704), nrow = 2, ncol = 2, byrow = T))
empty_raster <- setExtent(raster(nrows = 704, ncols = 1024), empty_raster_extent, keepres = F)
values(empty_raster) <- 0
panorama[[empty_xy_id[k]]] <- empty_raster
}
}
panorama_merged <- do.call(merge, panorama)
xray_brick_list[[j]] <- panorama_merged
}
}
message("Stitching complete. Creating x-ray brick...")
xray_brick <- do.call(brick, xray_brick_list)
names(xray_brick) <- unlist(xray_data)
message("...complete.")
message("Writing brick...")
if(!is.na(opt$o)){
dir.create(opt$o)
if(!is.na(opt$n)){
out_brick <- writeRaster(xray_brick, paste0(opt$o, "/", opt$n,"_brick.grd"), overwrite=TRUE, format="raster")
x <- writeRaster(xray_brick, paste0(opt$o, "/", opt$n,"_brick.tif"), overwrite=TRUE, format="GTiff",options=c("INTERLEAVE=BAND","COMPRESS=LZW"))
}else{
out_brick <- writeRaster(xray_brick, path = opt$o, "brick.grd", overwrite=TRUE, format="raster")
x <- writeRaster(xray_brick, path = opt$o, "brick.tif", overwrite=TRUE, format="GTiff",options=c("INTERLEAVE=BAND","COMPRESS=LZW"))
}
}else{
if(!is.na(opt$n)){
out_brick <- writeRaster(xray_brick, paste0(opt$n,"_brick.grd"), overwrite=TRUE, format="raster")
x <- writeRaster(xray_brick, paste0(opt$n,"_brick.tif"), overwrite=TRUE, format="GTiff",options=c("INTERLEAVE=BAND","COMPRESS=LZW"))
}else{
out_brick <- writeRaster(xray_brick, "brick.grd", overwrite=TRUE, format="raster")
x <- writeRaster(xray_brick, "brick.tif", overwrite=TRUE, format="GTiff",options=c("INTERLEAVE=BAND","COMPRESS=LZW"))
}
}
message("...complete.")
remove(list = c("x", "xray_brick_list", "xray_data", "empty_raster", "empty_raster_extent",
"i", "j", "k", "path", "positions", "xy_id",
"panorama_merged", "panorama", "empty_xy_id", "files", "directories"))
if(opt$p){
if(!is.na(opt$b)){
SEM_images <- list.files(opt$b, full.names = T, pattern = opt$m)
}else{
message("Please provide a file path for your SEM images to stitch. For help, see 'dataStitchR.R --help.'")
break
}
if(!is.na(opt$y)){
SEM_images <- SEM_images[!str_detect(SEM_images , opt$y)]
}
SEM_panorama <- list()
message("Stitching SEM images into panorama...")
for(i in 1:length(SEM_images)){
image <- SEM_images[i] %>% image_read() %>%
image_quantize(colorspace = 'gray') %>%
image_equalize()
temp_file <- tempfile()
image_write(image, path = temp_file, format = 'tiff')
image <- raster(temp_file)
image_extent <- extent(matrix(c(xy$x[i], xy$x[i] + 1024, xy$y[i], xy$y[i]+704), nrow = 2, ncol = 2, byrow = T))
image_raster <- setExtent(raster(nrows = 704, ncols = 1024), image_extent, keepres = F)
values(image_raster) <- values(image)
SEM_panorama[[i]] <- image_raster
}
SEM_panorama_merged <- do.call(merge, SEM_panorama)
message("...complete.")
remove(list = c("SEM_panorama", "SEM_images", "image", "image_extent", "image_raster"))
message("Generating plot...")
element_colors <- c("#FFFFFF", "#D9FFFF", "#CC80FF", "#C2FF00", "#FFB5B5", "#909090", "#3050F8",
"#FF0D0D", "#90E050", "#B3E3F5", "#AB5CF2", "#8AFF00", "#BFA6A6", "#F0C8A0",
"#FF8000", "#FFFF30", "#1FF01F", "#80D1E3", "#8F40D4", "#3DFF00", "#E6E6E6",
"#BFC2C7", "#A6A6AB", "#8A99C7", "#9C7AC7", "#E06633", "#F090A0", "#50D050",
"#C88033", "#7D80B0", "#C28F8F", "#668F8F", "#BD80E3", "#FFA100", "#A62929",
"#5CB8D1", "#702EB0", "#00FF00", "#94FFFF", "#94E0E0", "#73C2C9", "#54B5B5",
"#3B9E9E", "#248F8F", "#0A7D8C", "#006985", "#C0C0C0", "#FFD98F", "#A67573",
"#668080", "#9E63B5", "#D47A00", "#940094", "#429EB0", "#57178F", "#00C900",
"#70D4FF", "#FFFFC7", "#D9FFC7", "#C7FFC7", "#A3FFC7", "#8FFFC7", "#61FFC7",
"#45FFC7", "#30FFC7", "#1FFFC7", "#00FF9C", "#00E675", "#00D452", "#00BF38",
"#00AB24", "#4DC2FF", "#4DA6FF", "#2194D6", "#267DAB", "#266696", "#175487",
"#D0D0E0", "#FFD123", "#B8B8D0", "#A6544D", "#575961", "#9E4FB5", "#AB5C00",
"#754F45", "#428296", "#420066", "#007D00", "#70ABFA", "#00BAFF", "#00A1FF",
"#008FFF", "#0080FF", "#006BFF", "#545CF2", "#785CE3", "#8A4FE3", "#A136D4",
"#B31FD4", "#B31FBA", "#B30DA6", "#BD0D87", "#C70066", "#CC0059", "#D1004F",
"#D90045", "#E00038", "#E6002E", "#EB0026")
names(element_colors) <- c("H", "He", "Li", "Be", "B", "C", "N", "O", "F", "Ne", "Na", "Mg", "Al", "Si",
"P", "S", "Cl", "Ar", "K", "Ca", "Sc", "Ti", "V", "Cr", "Mn", "Fe", "Co", "Ni",
"Cu", "Zn", "Ga", "Ge", "As", "Se", "Br", "Kr", "Rb", "Sr", "Y", "Zr", "Nb", "Mo",
"Tc", "Ru", "Rh", "Pd", "Ag", "Cd", "In", "Sn", "Sb", "Te", "I", "Xe", "Cs", "Ba",
"La", "Ce", "Pr", "Nd", "Pm", "Sm", "Eu", "Gd", "Tb", "Dy", "Ho", "Er", "Tm", "Yb",
"Lu", "Hf", "Ta", "W", "Re", "Os", "Ir", "Pt", "Au", "Hg", "Tl", "Pb", "Bi", "Po",
"At", "Rn", "Fr", "Ra", "Ac", "Th", "Pa", "U", "Np", "Pu", "Am", "Cm", "Bk", "Cf",
"Es", "Fm", "Md", "No", "Lr", "Rf", "Db", "Sg", "Bh", "Hs", "Mt")
xray_frame <- as.data.frame(xray_brick, xy=TRUE)
xray_frame <- gather(xray_frame, element, value, colnames(xray_frame)[3]:colnames(xray_frame)[ncol(xray_frame)])
element_plotter<-function(coord_frame, brick, SEM_image, colors){
p <-rasterVis::gplot(SEM_image) +
geom_tile(aes(fill = value)) +
scale_fill_gradient(low = 'black', high = 'white') +
ggnewscale::new_scale_fill()
for(i in names(brick)){
message(paste0("Adding ", names(brick[[i]]), " to plot..."))
element_coords <- coord_frame %>%
filter(element == names(brick[[i]]) & value!=0)
p <- p+geom_raster(element_coords, mapping = aes(x, y, fill = element, alpha = value)) +
scale_fill_manual(values = colors) +
ggnewscale::new_scale_fill()
}
message("Writing plot...")
suppressWarnings(print(p +
coord_fixed() +
theme(axis.title = element_blank(),
axis.text = element_blank(),
legend.position = "none")))
}
element_plot <- element_plotter(xray_frame, xray_brick, SEM_panorama_merged, element_colors)
element_plot_legend <- data.frame(element = unique(xray_frame$element)) %>%
rownames_to_column() %>%
ggplot(aes(element, rowname, fill=element)) +
geom_bar(stat= "identity") +
scale_fill_manual(values = element_colors) +
guides(fill=guide_legend(nrow=2)) +
theme(legend.title = ggplot2::element_blank(),
legend.text = ggplot2::element_text(size = 8))
element_plot_with_legend <- plot_grid(
element_plot,
plot_grid(get_legend(element_plot_legend),
ncol = 1),
nrow = 2,
rel_heights = c(8,2)
)
if(!is.na(opt$n)){
if(!is.na(opt$o)){
pdf(paste0(opt$o, "/", opt$n, "_element_plot.pdf"),
width = 13.33,
height = 7.5)
}else{
pdf(paste0(opt$n, "_element_plot.pdf"),
width = 13.33,
height = 7.5)
}
}else{
if(!is.na(opt$o)){
pdf(paste0(opt$o, "/", "element_plot.pdf"),
width = 13.33,
height = 7.5)
}else{
pdf("element_plot.pdf",
width = 13.33,
height = 7.5)
}
}
print(element_plot_with_legend)
dev.off()
message("...complete.")
remove(list = ls())
}