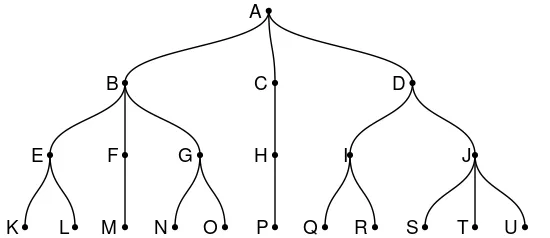
给定一个由包含符号边缘列表的数据框构建的有向树(igraph图形):
library(igraph)
library(ggraph)
#Create a symbolic edge list.
edgelist_df <- data.frame("from" = c("A", "A", "A", "B", "B", "B", "C", "D", "D", "E",
"E", "F", "G", "G", "H", "I", "I", "J", "J", "J"),
"to" = c("B", "C", "D", "E", "F", "G", "H", "I", "J", "K",
"L", "M", "N", "O", "P", "Q", "R", "S", "T", "U"))
#Create a directed tree from this edgelist.
graph <- graph_from_data_frame(d = edgelist_df, directed = TRUE)
绘制树形图。这里我使用ggraph包中的ggraph函数。
ggraph(graph = graph,
layout = 'dendrogram',
circular = FALSE) +
geom_edge_diagonal() +
geom_node_point() +
geom_node_text(aes(label = name),
angle = 0,
hjust = 1.5,
nudge_y = 0,
size = 5) +
theme_void()
- 如果节点为“B”,则属于以“B”为根节点的子树的所有叶节点为:“K”,“L”,“M”,“N”和“O”。
- 如果节点为“H”,则属于以“H”为根节点的子树的所有叶节点为:“P”。
- 如果节点为“A”,则属于以“A”为根节点(即原始树)的子树中的所有叶节点为:“K”,“L”,“M”,“N”,“O”,“P”,“Q”,“R”,“S”,“T”和“U”。