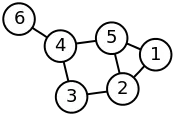
我试图学习如何创建网络图并从中生成稀疏矩阵。通过维基百科中的
以下是我用于完成此操作的代码,对于更大的网络,我感觉它非常低效。
Laplacian matrix 示例,我决定尝试使用 networkx 重新创建以下网络图:
如何高效转换 邻接矩阵 和 网络图 之间的关系?
例如,如果我有一个网络图,如何快速将其转换为邻接矩阵?如果我有一个邻接矩阵,如何高效地将其转换为网络图?以下是我用于完成此操作的代码,对于更大的网络,我感觉它非常低效。
#!/usr/bin/python
import networkx as nx
import numpy as np
import matplotlib.pyplot as plt
import scipy as sp
import pandas as pd
%matplotlib inline
#Adjacent matrix
adj_matrix = np.matrix([[0,1,0,0,1,0],[1,0,1,0,1,0],[0,1,0,1,0,0],[0,0,1,0,1,1],[1,1,0,1,0,0],[0,0,0,1,0,0]])
adj_sparse = sp.sparse.coo_matrix(adj_matrix, dtype=np.int8)
labels = range(1,7)
DF_adj = pd.DataFrame(adj_sparse.toarray(),index=labels,columns=labels)
print DF_adj
# 1 2 3 4 5 6
#1 0 1 0 0 1 0
#2 1 0 1 0 1 0
#3 0 1 0 1 0 0
#4 0 0 1 0 1 1
#5 1 1 0 1 0 0
#6 0 0 0 1 0 0
#Network graph
G = nx.Graph()
G.add_nodes_from(labels)
#Connect nodes
for i in range(DF_adj.shape[0]):
col_label = DF_adj.columns[i]
for j in range(DF_adj.shape[1]):
row_label = DF_adj.index[j]
node = DF_adj.iloc[i,j]
if node == 1:
G.add_edge(col_label,row_label)
#Draw graph
nx.draw(G,with_labels = True)
#DRAWN GRAPH MATCHES THE GRAPH FROM WIKI
#Recreate adjacency matrix
DF_re = pd.DataFrame(np.zeros([len(G.nodes()),len(G.nodes())]),index=G.nodes(),columns=G.nodes())
for col_label,row_label in G.edges():
DF_re.loc[col_label,row_label] = 1
DF_re.loc[row_label,col_label] = 1
print G.edges()
#[(1, 2), (1, 5), (2, 3), (2, 5), (3, 4), (4, 5), (4, 6)]
print DF_re
# 1 2 3 4 5 6
#1 0 1 0 0 1 0
#2 1 0 1 0 1 0
#3 0 1 0 1 0 0
#4 0 0 1 0 1 1
#5 1 1 0 1 0 0
#6 0 0 0 1 0 0