我正在尝试使用下面的代码通过Kmeans分割肺部CT图像:
def process_mask(mask):
convex_mask = np.copy(mask)
for i_layer in range(convex_mask.shape[0]):
mask1 = np.ascontiguousarray(mask[i_layer])
if np.sum(mask1)>0:
mask2 = convex_hull_image(mask1)
if np.sum(mask2)>2*np.sum(mask1):
mask2 = mask1
else:
mask2 = mask1
convex_mask[i_layer] = mask2
struct = generate_binary_structure(3,1)
dilatedMask = binary_dilation(convex_mask,structure=struct,iterations=10)
return dilatedMask
def lumTrans(img):
lungwin = np.array([-1200.,600.])
newimg = (img-lungwin[0])/(lungwin[1]-lungwin[0])
newimg[newimg<0]=0
newimg[newimg>1]=1
newimg = (newimg*255).astype('uint8')
return newimg
def lungSeg(imgs_to_process,output,name):
if os.path.exists(output+'/'+name+'_clean.npy') : return
imgs_to_process = Image.open(imgs_to_process)
img_to_save = imgs_to_process.copy()
img_to_save = np.asarray(img_to_save).astype('uint8')
imgs_to_process = lumTrans(imgs_to_process)
imgs_to_process = np.expand_dims(imgs_to_process, axis=0)
x,y,z = imgs_to_process.shape
img_array = imgs_to_process.copy()
A1 = int(y/(512./100))
A2 = int(y/(512./400))
A3 = int(y/(512./475))
A4 = int(y/(512./40))
A5 = int(y/(512./470))
for i in range(len(imgs_to_process)):
img = imgs_to_process[i]
print(img.shape)
x,y = img.shape
#Standardize the pixel values
allmean = np.mean(img)
allstd = np.std(img)
img = img-allmean
img = img/allstd
# Find the average pixel value near the lungs
# to renormalize washed out images
middle = img[A1:A2,A1:A2]
mean = np.mean(middle)
max = np.max(img)
min = np.min(img)
kmeans = KMeans(n_clusters=2).fit(np.reshape(middle,[np.prod(middle.shape),1]))
centers = sorted(kmeans.cluster_centers_.flatten())
threshold = np.mean(centers)
thresh_img = np.where(img<threshold,1.0,0.0) # threshold the image
eroded = morphology.erosion(thresh_img,np.ones([4,4]))
dilation = morphology.dilation(eroded,np.ones([10,10]))
labels = measure.label(dilation)
label_vals = np.unique(labels)
regions = measure.regionprops(labels)
good_labels = []
for prop in regions:
B = prop.bbox
if B[2]-B[0]<A3 and B[3]-B[1]<A3 and B[0]>A4 and B[2]<A5:
good_labels.append(prop.label)
mask = np.ndarray([x,y],dtype=np.int8)
mask[:] = 0
for N in good_labels:
mask = mask + np.where(labels==N,1,0)
mask = morphology.dilation(mask,np.ones([10,10])) # one last dilation
imgs_to_process[i] = mask
m1 = imgs_to_process
convex_mask = m1
dm1 = process_mask(m1)
dilatedMask = dm1
Mask = m1
extramask = dilatedMask ^ Mask
bone_thresh = 180
pad_value = 0
img_array[np.isnan(img_array)]=-2000
sliceim = img_array
sliceim = sliceim*dilatedMask+pad_value*(1-dilatedMask).astype('uint8')
bones = sliceim*extramask>bone_thresh
sliceim[bones] = pad_value
x,y,z = sliceim.shape
if not os.path.exists(output):
os.makedirs(output)
img_to_save[sliceim.squeeze()==0] = 0
im = Image.fromarray(img_to_save)
im.save(output + name + '.png', 'PNG')
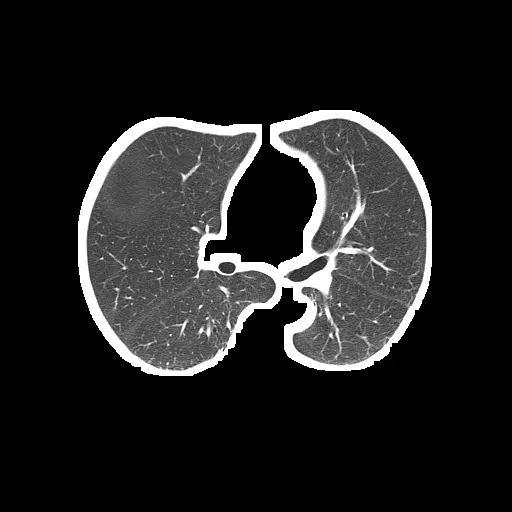
问题是分割后的肺部仍然包含像这样的白色边框:
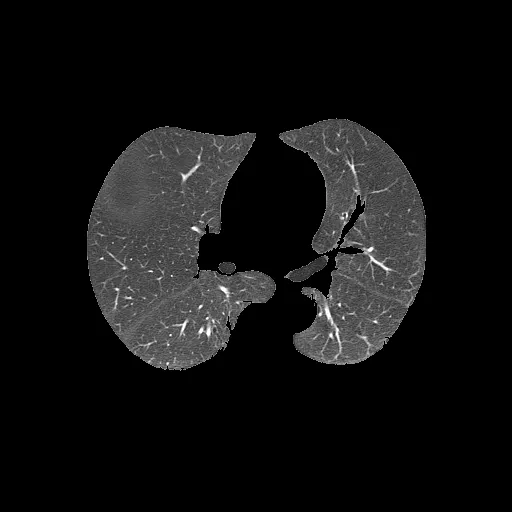
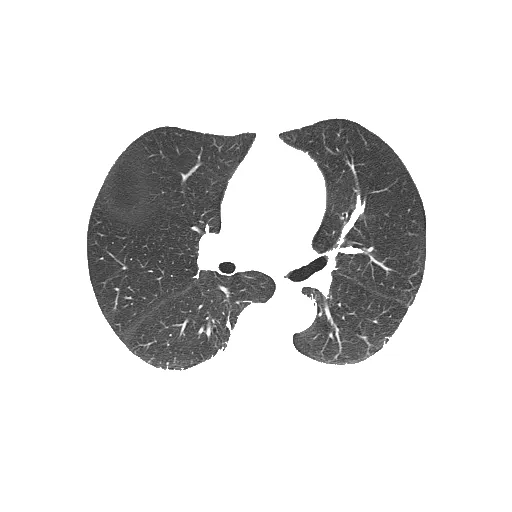
分割后的肺部(输出):
未分割的肺部(输入):
完整代码可以在Google Colab笔记本中找到。code。
数据集的示例在这里。
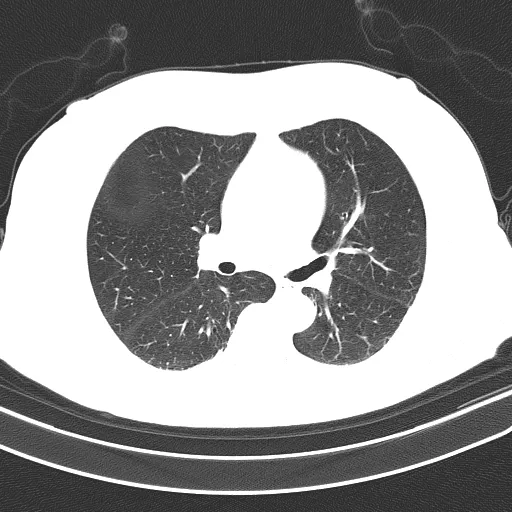
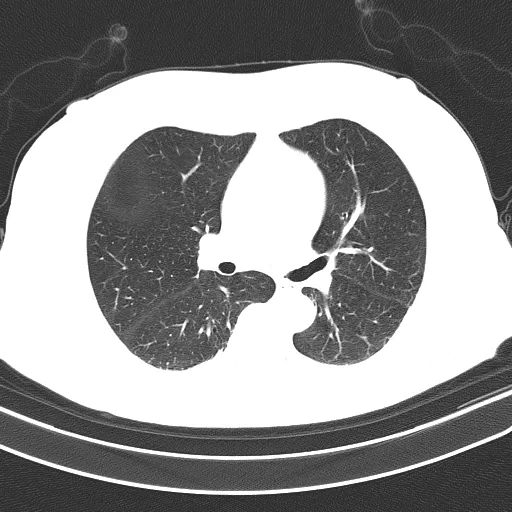
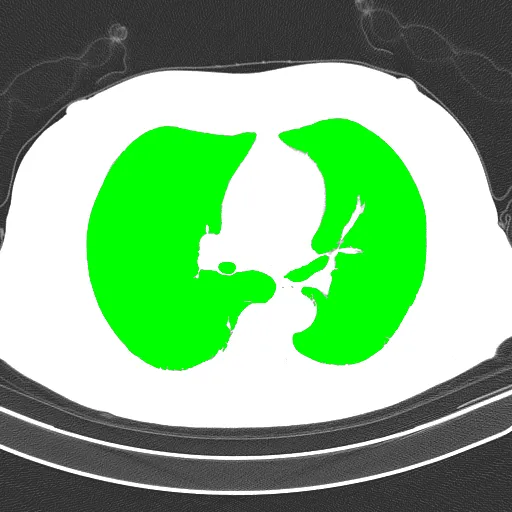
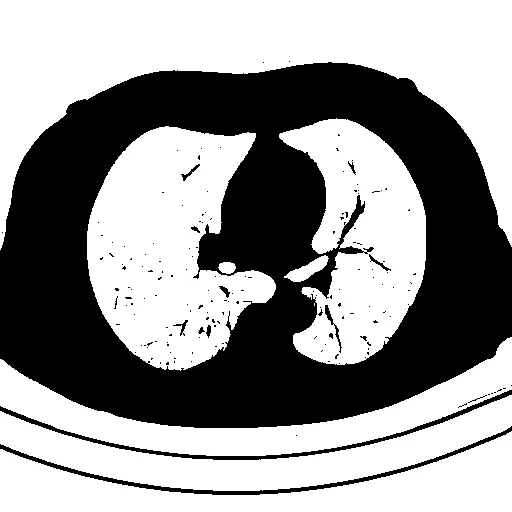 检测到需要提取的对象会用绿色标出
检测到需要提取的对象会用绿色标出