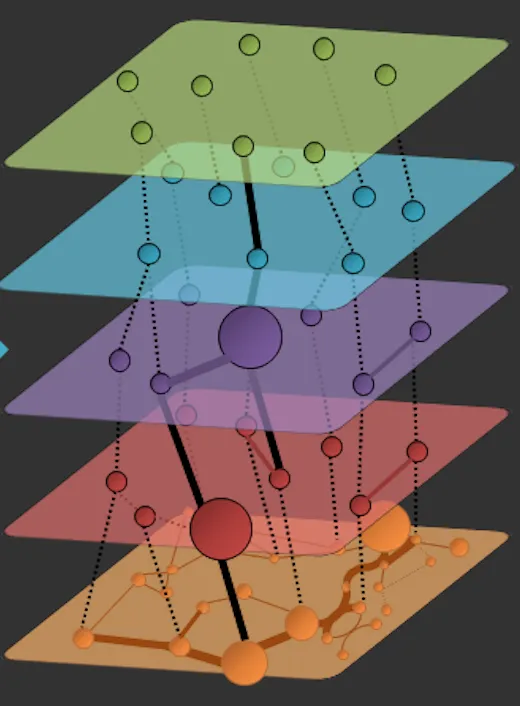
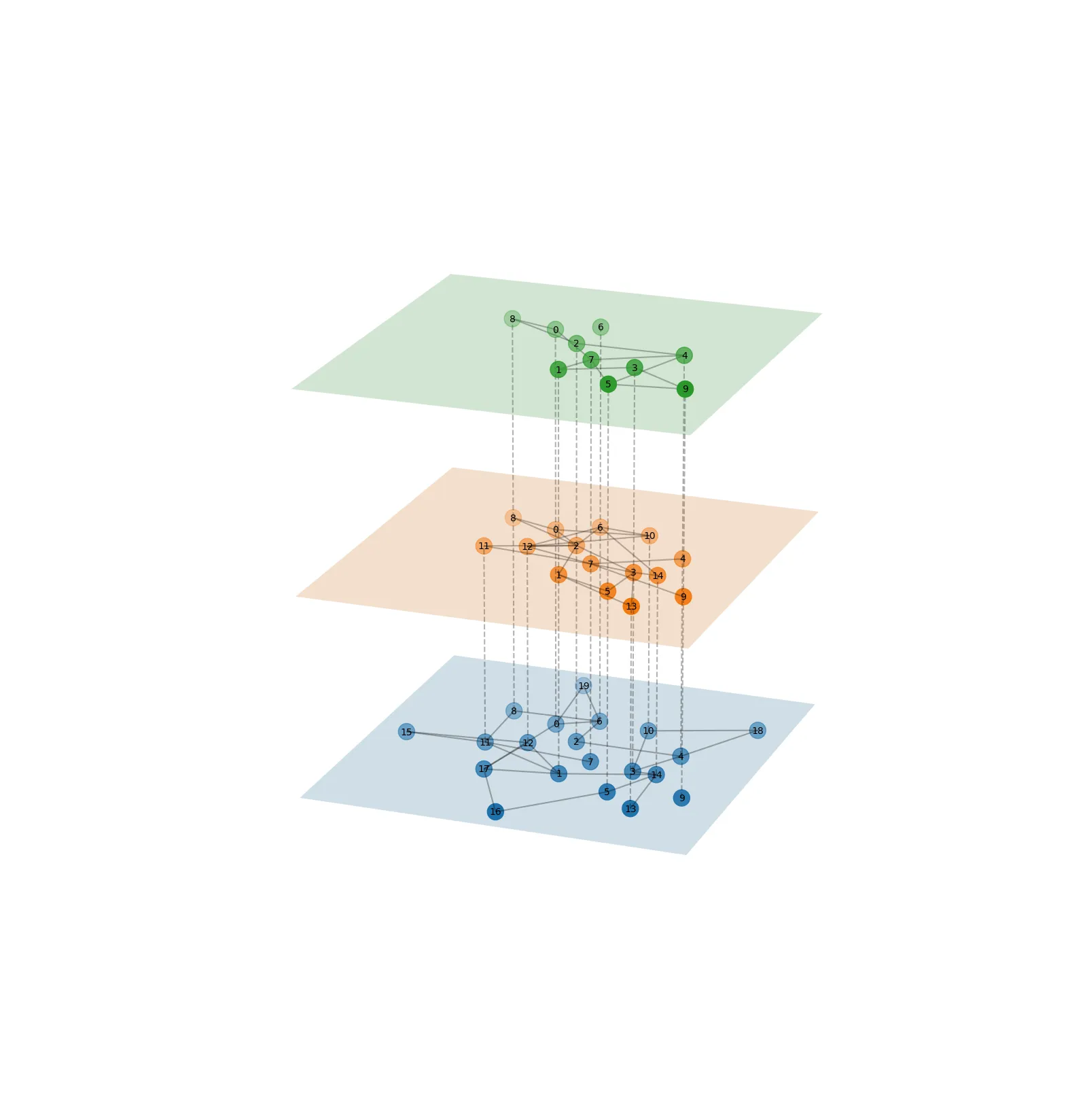
networkx 目前没有内置支持分层布局的功能,更不用说像示例中展示的可视化。因此我们需要自己动手实现。
下面的实现 LayeredNetworkGraph 假设你有一个表示不同层的图形列表 [g1, g2, ..., gn]。在一层内,相应的(子)图定义了连接性。在层之间,后续层中的节点如果具有相同的节点ID,则相互连接。
由于没有(据我所知)能够在三维空间中计算具有层内节点平面约束的节点位置的布局函数,因此我们使用一个小技巧:在所有层之间创建图形组合,计算二维位置,然后将这些位置应用于所有层中的节点。可以使用真正的力导向布局来计算带有平面约束的布局,但这将是很多工作,而且由于您的示例仅使用了shell布局(不受影响),因此我没有费心。在许多情况下,差异会很小。
如果您想更改可视化方面的某些内容(如大小、宽度、颜色),请查看 draw 方法。您可能需要进行的大多数更改都可以在那里完成。
"""
Plot multi-graphs in 3D.
"""
import numpy as np
import matplotlib.pyplot as plt
import networkx as nx
from mpl_toolkits.mplot3d import Axes3D
from mpl_toolkits.mplot3d.art3d import Line3DCollection
class LayeredNetworkGraph(object):
def __init__(self, graphs, node_labels=None, layout=nx.spring_layout, ax=None):
"""Given an ordered list of graphs [g1, g2, ..., gn] that represent
different layers in a multi-layer network, plot the network in
3D with the different layers separated along the z-axis.
Within a layer, the corresponding graph defines the connectivity.
Between layers, nodes in subsequent layers are connected if
they have the same node ID.
Arguments:
----------
graphs : list of networkx.Graph objects
List of graphs, one for each layer.
node_labels : dict node ID : str label or None (default None)
Dictionary mapping nodes to labels.
If None is provided, nodes are not labelled.
layout_func : function handle (default networkx.spring_layout)
Function used to compute the layout.
ax : mpl_toolkits.mplot3d.Axes3d instance or None (default None)
The axis to plot to. If None is given, a new figure and a new axis are created.
"""
self.graphs = graphs
self.total_layers = len(graphs)
self.node_labels = node_labels
self.layout = layout
if ax:
self.ax = ax
else:
fig = plt.figure()
self.ax = fig.add_subplot(111, projection='3d')
self.get_nodes()
self.get_edges_within_layers()
self.get_edges_between_layers()
self.get_node_positions()
self.draw()
def get_nodes(self):
"""Construct an internal representation of nodes with the format (node ID, layer)."""
self.nodes = []
for z, g in enumerate(self.graphs):
self.nodes.extend([(node, z) for node in g.nodes()])
def get_edges_within_layers(self):
"""Remap edges in the individual layers to the internal representations of the node IDs."""
self.edges_within_layers = []
for z, g in enumerate(self.graphs):
self.edges_within_layers.extend([((source, z), (target, z)) for source, target in g.edges()])
def get_edges_between_layers(self):
"""Determine edges between layers. Nodes in subsequent layers are
thought to be connected if they have the same ID."""
self.edges_between_layers = []
for z1, g in enumerate(self.graphs[:-1]):
z2 = z1 + 1
h = self.graphs[z2]
shared_nodes = set(g.nodes()) & set(h.nodes())
self.edges_between_layers.extend([((node, z1), (node, z2)) for node in shared_nodes])
def get_node_positions(self, *args, **kwargs):
"""Get the node positions in the layered layout."""
composition = self.graphs[0]
for h in self.graphs[1:]:
composition = nx.compose(composition, h)
pos = self.layout(composition, *args, **kwargs)
self.node_positions = dict()
for z, g in enumerate(self.graphs):
self.node_positions.update({(node, z) : (*pos[node], z) for node in g.nodes()})
def draw_nodes(self, nodes, *args, **kwargs):
x, y, z = zip(*[self.node_positions[node] for node in nodes])
self.ax.scatter(x, y, z, *args, **kwargs)
def draw_edges(self, edges, *args, **kwargs):
segments = [(self.node_positions[source], self.node_positions[target]) for source, target in edges]
line_collection = Line3DCollection(segments, *args, **kwargs)
self.ax.add_collection3d(line_collection)
def get_extent(self, pad=0.1):
xyz = np.array(list(self.node_positions.values()))
xmin, ymin, _ = np.min(xyz, axis=0)
xmax, ymax, _ = np.max(xyz, axis=0)
dx = xmax - xmin
dy = ymax - ymin
return (xmin - pad * dx, xmax + pad * dx), \
(ymin - pad * dy, ymax + pad * dy)
def draw_plane(self, z, *args, **kwargs):
(xmin, xmax), (ymin, ymax) = self.get_extent(pad=0.1)
u = np.linspace(xmin, xmax, 10)
v = np.linspace(ymin, ymax, 10)
U, V = np.meshgrid(u ,v)
W = z * np.ones_like(U)
self.ax.plot_surface(U, V, W, *args, **kwargs)
def draw_node_labels(self, node_labels, *args, **kwargs):
for node, z in self.nodes:
if node in node_labels:
ax.text(*self.node_positions[(node, z)], node_labels[node], *args, **kwargs)
def draw(self):
self.draw_edges(self.edges_within_layers, color='k', alpha=0.3, linestyle='-', zorder=2)
self.draw_edges(self.edges_between_layers, color='k', alpha=0.3, linestyle='--', zorder=2)
for z in range(self.total_layers):
self.draw_plane(z, alpha=0.2, zorder=1)
self.draw_nodes([node for node in self.nodes if node[1]==z], s=300, zorder=3)
if self.node_labels:
self.draw_node_labels(self.node_labels,
horizontalalignment='center',
verticalalignment='center',
zorder=100)
if __name__ == '__main__':
n = 5
g = nx.erdos_renyi_graph(4*n, p=0.1)
h = nx.erdos_renyi_graph(3*n, p=0.2)
i = nx.erdos_renyi_graph(2*n, p=0.4)
node_labels = {nn : str(nn) for nn in range(4*n)}
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
LayeredNetworkGraph([g, h, i], node_labels=node_labels, ax=ax, layout=nx.spring_layout)
ax.set_axis_off()
plt.show()