我正在尝试比较一些循环神经网络,但只有LSTM出现了问题,我不知道为什么。
我正在使用相同的代码/数据集训练LSTM、SimpleRNN和GRU。对于所有模型,损失都会正常降低。但是对于LSTM,在某个点(损失约为0.4)之后,损失直接降至10e-8。如果我尝试预测输出,只会得到NaN。
以下是代码:
这是使用相同输入的GRU和LSTM的输出:
我正在使用相同的代码/数据集训练LSTM、SimpleRNN和GRU。对于所有模型,损失都会正常降低。但是对于LSTM,在某个点(损失约为0.4)之后,损失直接降至10e-8。如果我尝试预测输出,只会得到NaN。
以下是代码:
nb_unit = 7
inp_shape = (maxlen, 7)
loss_ = "categorical_crossentropy"
metrics_ = "categorical_crossentropy"
optimizer_ = "Nadam"
nb_epoch = 250
batch_size = 64
model = Sequential()
model.add(LSTM( units=nb_unit,
input_shape=inp_shape,
return_sequences=True,
activation='softmax')) # I just change the cell name
model.compile(loss=loss_,
optimizer=optimizer_,
metrics=[metrics_])
checkpoint = ModelCheckpoint("lstm_simple.h5",
monitor=loss_,
verbose=1,
save_best_only=True,
save_weights_only=False,
mode='auto',
period=1)
early = EarlyStopping( monitor='loss',
min_delta=0,
patience=10,
verbose=1,
mode='auto')
history = model.fit(X_train, y_train,
validation_data=(X_test, y_test),
epochs=nb_epoch,
batch_size=batch_size,
verbose=2,
callbacks = [checkpoint, early])
这是使用相同输入的GRU和LSTM的输出:
Input :
[[[1 0 0 0 0 0 0]
[0 1 0 0 0 0 0]
[0 0 0 1 0 0 0]
[0 0 0 1 0 0 0]
[0 1 0 0 0 0 0]
[0 0 0 0 0 1 0]
[0 0 0 0 1 0 0]
[0 0 0 1 0 0 0]
[0 0 0 0 0 1 0]
[0 0 0 0 1 0 0]
[0 0 0 1 0 0 0]
[0 1 0 0 0 0 0]
[0 0 0 0 0 1 0]
[0 0 0 0 1 0 0]
[0 0 0 1 0 0 0]
[0 0 0 0 0 1 0]
[0 0 0 0 0 1 0]
[0 0 0 0 0 0 0]
[0 0 0 0 0 0 0]
[0 0 0 0 0 0 0]]]
LSTM predicts :
[[[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]
[ nan nan nan nan nan nan nan]]]
GRU predicts :
[[[ 0. 0.54 0. 0. 0.407 0. 0. ]
[ 0. 0.005 0.66 0.314 0. 0. 0.001]
[ 0. 0.001 0.032 0.957 0. 0.004 0. ]
[ 0. 0.628 0. 0. 0. 0.372 0. ]
[ 0. 0.555 0. 0. 0. 0.372 0. ]
[ 0. 0. 0. 0. 0.996 0.319 0. ]
[ 0. 0. 0.167 0.55 0. 0. 0. ]
[ 0. 0.486 0. 0.002 0. 0.51 0. ]
[ 0. 0.001 0. 0. 0.992 0.499 0. ]
[ 0. 0. 0.301 0.55 0. 0. 0. ]
[ 0. 0.396 0.001 0.007 0. 0.592 0. ]
[ 0. 0.689 0. 0. 0. 0.592 0. ]
[ 0. 0.001 0. 0. 0.997 0.592 0. ]
[ 0. 0. 0.37 0.55 0. 0. 0. ]
[ 0. 0.327 0.003 0.025 0. 0.599 0. ]
[ 0. 0.001 0. 0. 0.967 0.599 0.002]
[ 0. 0. 0. 0. 0. 0.002 0.874]
[ 0.004 0.076 0.128 0.337 0.02 0.069 0.378]
[ 0.006 0.379 0.047 0.113 0.029 0.284 0.193]
[ 0.006 0.469 0.001 0.037 0.13 0.295 0.193]]]
针对损失,您可以在fit()历史记录的最后几行中找到以下内容:
Epoch 116/250
Epoch 00116: categorical_crossentropy did not improve
- 2s - loss: 0.3774 - categorical_crossentropy: 0.3774 - val_loss: 0.3945 - val_categorical_crossentropy: 0.3945
Epoch 117/250
Epoch 00117: categorical_crossentropy improved from 0.37673 to 0.08198, saving model to lstm_simple.h5
- 2s - loss: 0.0820 - categorical_crossentropy: 0.0820 - val_loss: 7.8743e-08 - val_categorical_crossentropy: 7.8743e-08
Epoch 118/250
Epoch 00118: categorical_crossentropy improved from 0.08198 to 0.00000, saving model to lstm_simple.h5
- 2s - loss: 7.5460e-08 - categorical_crossentropy: 7.5460e-08 - val_loss: 7.8743e-08 - val_categorical_crossentropy: 7.8743e-08
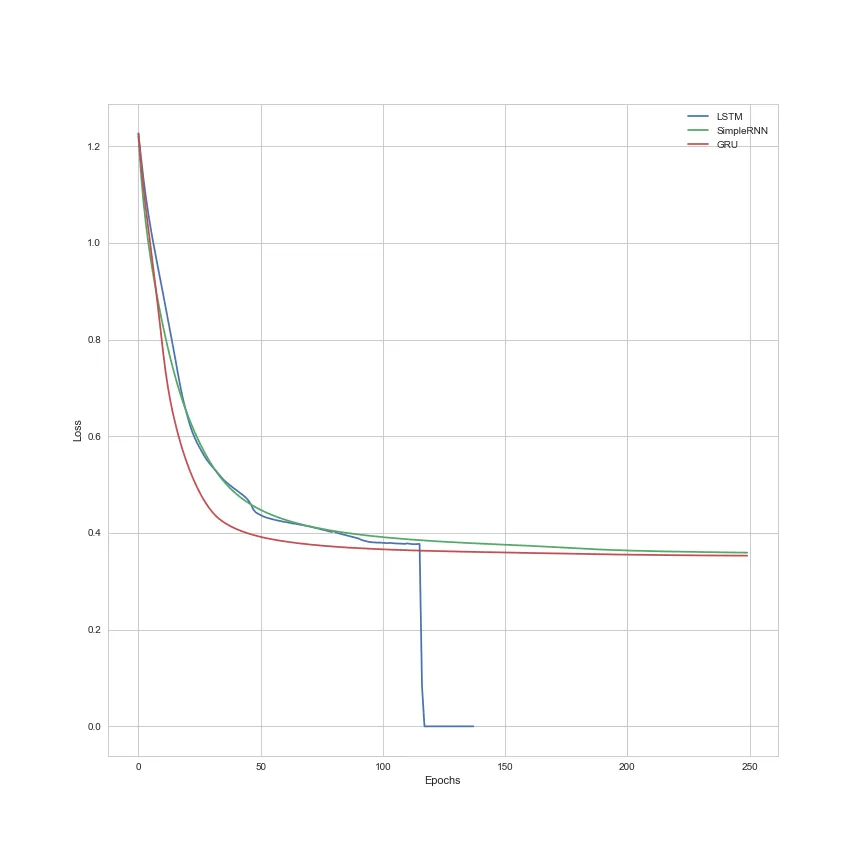
或者基于Epochs的损失演变。
我之前尝试过不使用Softmax和使用MSE作为损失函数,但没有出现任何错误。
如果需要,您可以在Github上找到笔记本和生成数据集的脚本(https://github.com/Coni63/SO/blob/master/Reber.ipynb)。
非常感谢您的支持, 问候, Nicolas
编辑1:
根本原因似乎是Softmax函数消失了。如果在崩溃之前停止并显示每个时间步长的softmax总和,则会得到:
LSTM :
[[ 0.112]
[ 0.008]
[ 0.379]
[ 0.04 ]
[ 0.001]
[ 0.104]
[ 0.021]
[ 0. ]
[ 0.104]
[ 0.343]
[ 0.012]
[ 0. ]
[ 0.23 ]
[ 0.13 ]
[ 0.147]
[ 0.145]
[ 0.152]
[ 0.157]
[ 0.163]
[ 0.169]]
GRU :
[[ 0.974]
[ 0.807]
[ 0.719]
[ 1.184]
[ 0.944]
[ 0.999]
[ 1.426]
[ 0.957]
[ 0.999]
[ 1.212]
[ 1.52 ]
[ 0.954]
[ 0.42 ]
[ 0.83 ]
[ 0.903]
[ 0.944]
[ 0.976]
[ 1.005]
[ 1.022]
[ 1.029]]
当Softmax为0时,下一步将尝试除以0。现在我不知道如何解决。
return_sequences设置为True?当我把它设置为False,只使用最后一个时间步时,会出现错误。 - Tommy Wolfheartreturn_sequences=False,那么我得到的输出维度是batch_size x num_features,而如果是True,则会变成batch_size x sequence_length x features。所以似乎categorical_crossentropy或其他东西不喜欢这样。无论如何,你可能需要更多上下文才能提供帮助,所以没关系。 - Tommy Wolfheart