我在使用Python中的NetworkX。给定任何无向且无权重的图,我想要循环遍历所有节点。对于每个节点,我希望有概率p添加一个随机边和/或删除一个现有的随机边。是否有简单的方法来实现这一点?非常感谢!
2个回答
12
在networkx中创建新的随机边
让我们先建立一个测试图:
import networkx as nx
import random
import matplotlib.pyplot as plt
graph = nx.Graph()
graph.add_edges_from([(1,3), (3,5), (2,4)])
nx.draw(graph, with_labels=True)
plt.show()
现在我们可以从图中的非边列表中随机选择一条边。目前还不清楚您所提到的概率是什么。由于您添加了一条评论,说明您想要使用random.choice,因此我将坚持这种方式。
def random_edge(graph, del_orig=True):
'''
Create a new random edge and delete one of its current edge if del_orig is True.
:param graph: networkx graph
:param del_orig: bool
:return: networkx graph
'''
edges = list(graph.edges)
nonedges = list(nx.non_edges(graph))
# random edge choice
chosen_edge = random.choice(edges)
chosen_nonedge = random.choice([x for x in nonedges if chosen_edge[0] == x[0]])
if del_orig:
# delete chosen edge
graph.remove_edge(chosen_edge[0], chosen_edge[1])
# add new edge
graph.add_edge(chosen_nonedge[0], chosen_nonedge[1])
return graph
使用示例:
new_graph = random_edge(graph, del_orig=True)
nx.draw(new_graph, with_labels=True)
plt.show()
如果需要,我们仍然可以在random.choice中添加边缘的概率分布(例如使用numpy.random.choice())。
- michaelg
2
给定一个节点i,为了避免添加重复的边,您需要知道(1)来自i的哪些边已经存在,然后计算(2)从i不存在的候选边集。对于删除操作,您已经在注释中定义了一种方法-它仅基于(1)。下面是一个函数,它将基于列表理解提供一轮随机添加和删除。
def add_and_remove_edges(G, p_new_connection, p_remove_connection):
'''
for each node,
add a new connection to random other node, with prob p_new_connection,
remove a connection, with prob p_remove_connection
operates on G in-place
'''
new_edges = []
rem_edges = []
for node in G.nodes():
# find the other nodes this one is connected to
connected = [to for (fr, to) in G.edges(node)]
# and find the remainder of nodes, which are candidates for new edges
unconnected = [n for n in G.nodes() if not n in connected]
# probabilistically add a random edge
if len(unconnected): # only try if new edge is possible
if random.random() < p_new_connection:
new = random.choice(unconnected)
G.add_edge(node, new)
print "\tnew edge:\t {} -- {}".format(node, new)
new_edges.append( (node, new) )
# book-keeping, in case both add and remove done in same cycle
unconnected.remove(new)
connected.append(new)
# probabilistically remove a random edge
if len(connected): # only try if an edge exists to remove
if random.random() < p_remove_connection:
remove = random.choice(connected)
G.remove_edge(node, remove)
print "\tedge removed:\t {} -- {}".format(node, remove)
rem_edges.append( (node, remove) )
# book-keeping, in case lists are important later?
connected.remove(remove)
unconnected.append(remove)
return rem_edges, new_edges
要看到此功能的效果,请执行以下操作:
import networkx as nx
import random
import matplotlib.pyplot as plt
p_new_connection = 0.1
p_remove_connection = 0.1
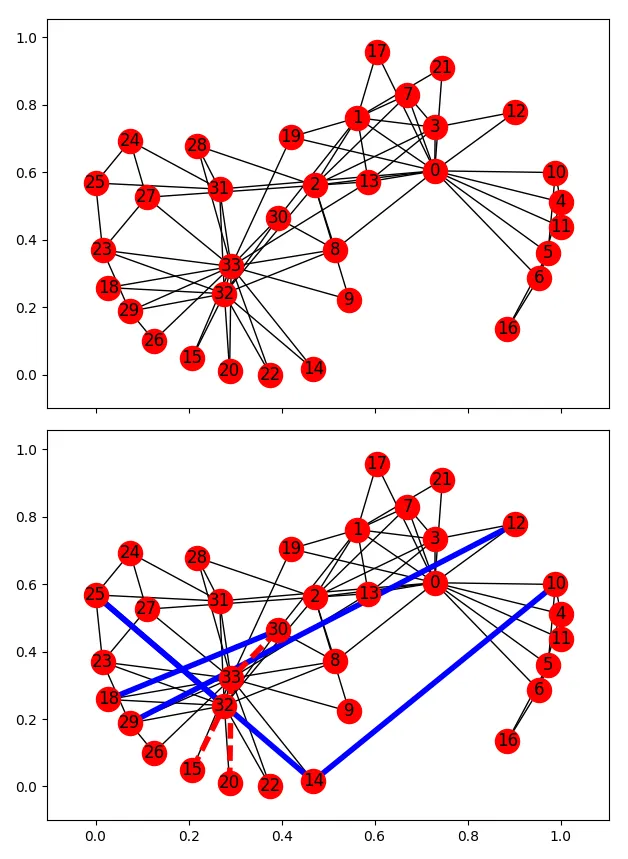
G = nx.karate_club_graph() # sample graph (undirected, unweighted)
# show original
plt.figure(1); plt.clf()
fig, ax = plt.subplots(2,1, num=1, sharex=True, sharey=True)
pos = nx.spring_layout(G)
nx.draw_networkx(G, pos=pos, ax=ax[0])
# now apply one round of changes
rem_edges, new_edges = add_and_remove_edges(G, p_new_connection, p_remove_connection)
# and draw new version and highlight changes
nx.draw_networkx(G, pos=pos, ax=ax[1])
nx.draw_networkx_edges(G, pos=pos, ax=ax[1], edgelist=new_edges,
edge_color='b', width=4)
# note: to highlight edges that were removed, add them back in;
# This is obviously just for display!
G.add_edges_from(rem_edges)
nx.draw_networkx_edges(G, pos=pos, ax=ax[1], edgelist=rem_edges,
edge_color='r', style='dashed', width=4)
G.remove_edges_from(rem_edges)
plt.show()
请注意,您也可以使用邻接矩阵进行类似操作,
A = nx.adjacency_matrix(G).todense() (它是一个numpy矩阵,因此像A[i,:] .nonzero() 这样的操作会很有用)。如果您有极大的网络,则可能更有效。
- Bonlenfum
网页内容由stack overflow 提供, 点击上面的可以查看英文原文,
原文链接
原文链接