另一个选择是使用
==。
. <- unique(df$x)
cbind(df, +do.call(cbind, lapply(setNames(., .), `==`, df$x)))
或者使用sapply在一行中完成。
cbind(df, +sapply(unique(df$x), `==`, df$x))
或者使用contrasts并将其与df$x匹配。
. <- contrasts(as.factor(df$x), FALSE)
cbind(df, .[match(df$x, rownames(.)),])
在矩阵中进行索引,即使用
matrix。
. <- unique(df$x)
i <- match(df$x, .)
nc <- length(.)
nr <- length(i)
cbind(df, matrix(`[<-`(integer(nc * nr), 1:nr + nr * (i - 1), 1), nr, nc,
dimnames=list(NULL, .)))
或者使用
outer。
. <- unique(df$x)
cbind(df, +outer(df$x, setNames(., .), `==`))
或者使用
rep和`matrix`。
. <- unique(df$x)
n <- nrow(df)
cbind(df, +matrix(df$x == rep(., each=n), n, dimnames=list(NULL, .)))
对于更多变量 x 中的代码,而不仅仅是 e.g. LETTERS,一些可用的方法进行基准测试。
set.seed(42)
df <- data.frame(n = seq(1:1000000), x = sample(LETTERS, 1000000, replace = T))
library(nnet)
library(dplyr)
microbenchmark::microbenchmark(times = 10L, setup = gc(FALSE), control=list(order="block")
, "nnet" = df %>% cbind(class.ind(.$x) == 1) %>%
mutate(across(-c(n, x), ~.*1))
, "contrasts" = {. <- contrasts(as.factor(df$x), FALSE)
cbind(df, .[match(df$x, rownames(.)),])}
, "==" = {. <- unique(df$x)
cbind(df, +do.call(cbind, lapply(setNames(., .), `==`, df$x)))}
, "==Sapply" = cbind(df, +sapply(unique(df$x), `==`, df$x))
, "matrix" = {. <- unique(df$x)
i <- match(df$x, .)
nc <- length(.)
nr <- length(i)
cbind(df, matrix(`[<-`(integer(nc * nr), 1:nr + nr * (i - 1), 1), nr, nc,
dimnames=list(NULL, .)))}
, "outer" = {. <- unique(df$x)
cbind(df, +outer(df$x, setNames(., .), `==`))}
, "rep" = {. <- unique(df$x)
n <- nrow(df)
cbind(df, +matrix(df$x == rep(., each=n), n, dimnames=list(NULL, .)))}
)
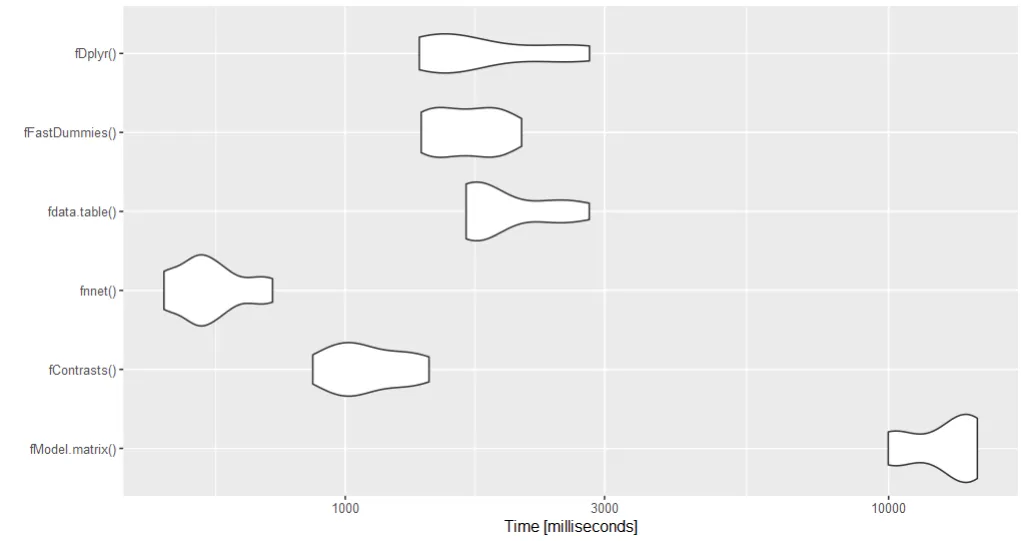
结果
Unit: milliseconds
expr min lq mean median uq max neval
nnet 208.6898 220.2304 326.2210 305.5752 386.3385 541.0621 10
contrasts 1110.0123 1168.7651 1263.5357 1216.1403 1357.0532 1514.4411 10
== 146.2217 156.8141 208.2733 185.1860 275.3909 278.8497 10
==Sapply 290.0458 291.4543 301.3010 295.0557 298.0274 358.0531 10
matrix 302.9993 304.8305 312.9748 306.8981 310.0781 363.0773 10
outer 524.5230 583.5224 603.3300 586.3054 595.4086 807.0260 10
rep 276.2110 285.3983 389.8187 434.2754 435.8607 442.3403 10