library(rpart)
train <- data.frame(ClaimID = c(1,2,3,4,5,6,7,8,9,10),
RearEnd = c(TRUE, TRUE, TRUE, FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, FALSE),
Whiplash = c(TRUE, TRUE, TRUE, TRUE, TRUE, FALSE, FALSE, FALSE, FALSE, TRUE),
Activity = factor(c("active", "very active", "very active", "inactive", "very inactive", "inactive", "very inactive", "active", "active", "very active"),
levels=c("very inactive", "inactive", "active", "very active"),
ordered=TRUE),
Fraud = c(FALSE, TRUE, TRUE, FALSE, FALSE, TRUE, TRUE, FALSE, FALSE, TRUE))
mytree <- rpart(Fraud ~ RearEnd + Whiplash + Activity, data = train, method = "class", minsplit = 2, minbucket = 1, cp=-1)
prp(mytree, type = 4, extra = 101, leaf.round = 0, fallen.leaves = TRUE,
varlen = 0, tweak = 1.2)
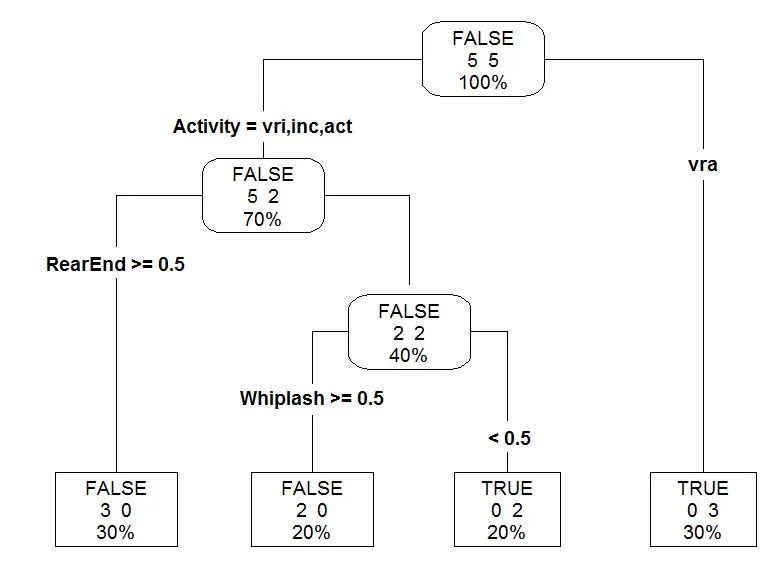
然后通过使用printcp我可以查看交叉验证结果。
> printcp(mytree)
Classification tree:
rpart(formula = Fraud ~ RearEnd + Whiplash + Activity, data = train,
method = "class", minsplit = 2, minbucket = 1, cp = -1)
Variables actually used in tree construction:
[1] Activity RearEnd Whiplash
Root node error: 5/10 = 0.5
n= 10
CP nsplit rel error xerror xstd
1 0.6 0 1.0 2.0 0.0
2 0.2 1 0.4 0.4 0.3
3 -1.0 3 0.0 0.4 0.3
因此,根节点误差为0.5,从我的理解来看,这是分类错误率。但我在计算灵敏度(真正例的比例)和特异性(真负例的比例)方面遇到了麻烦。我如何基于
rpart输出计算这些值?(上面的示例来自http://gormanalysis.com/decision-trees-in-r-using-rpart/)