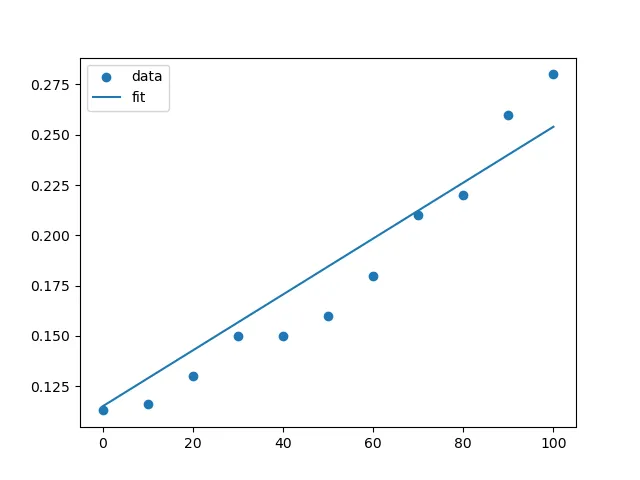
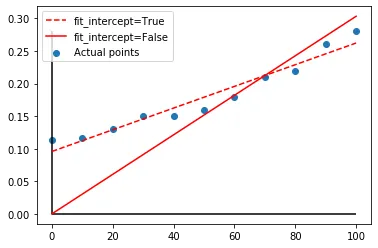
我希望进行一次线性回归,其中y截距被强制设为0.115。这是我尝试的代码。我设置fit_intercept=True以获得一个非零的y截距,但我能将其设置为一个值吗?
另外,我如何绘制最佳拟合直线,而不是连接每个点的直线?
提前感谢。
import numpy as np
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from sklearn.metrics import r2_score
from sklearn.linear_model import LinearRegression
x=np.array([0, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100]).reshape(-1,1)
y=np.array([0.113, 0.116, 0.130, 0.150, 0.150, 0.160, 0.180, 0.210, 0.220, 0.260, 0.280])
regression=LinearRegression(fit_intercept=True).fit(x,y)
r_sq=round(regression.score(x,y),4)
m=round(regression.coef_[0],4)
b=round(regression.intercept_,4)
print("r_sq:", r_sq,"m:",m,"b:",b)
plt.figure()
plt.scatter(x,y)
plt.title('A')
plt.ylabel('X')
plt.xlabel('Y')
plt.plot(x,y,'r--',label='measured')
plt.legend(loc='best')