2023年2月更新
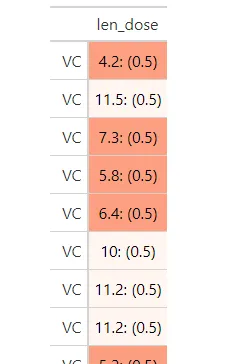
gt包现已添加了基于其他列进行着色的选项- data_color()现已获得taregt_columns参数。因此,这变得更加简单:
library(gt)
library(dplyr)
ToothGrowth %>%
mutate(len_dose = glue('{len}: ({dose})')) %>%
gt(rowname_col = 'supp') %>%
cols_hide(c(len, dose)) %>%
data_color(columns = "dose", target_columns = "len_dose",
palette = "ggsci::green_material")
已过时
我遇到了同样的问题,并调整了 gt::data_color 函数以接受单独的源和目标列 - 这样,下面的代码应该可以生成您所需的输出。
my_data_color <- function (data, SOURCE_columns, TARGET_columns, colors, alpha = NULL, apply_to = c("fill",
"text"), autocolor_text = TRUE)
{
stop_if_not_gt(data = data)
apply_to <- match.arg(apply_to)
colors <- rlang::enquo(colors)
data_tbl <- dt_data_get(data = data)
colors <- rlang::eval_tidy(colors, data_tbl)
resolved_source_columns <- resolve_cols_c(expr = {
{
SOURCE_columns
}
}, data = data)
resolved_target_columns <- resolve_cols_c(expr = {
{
TARGET_columns
}
}, data = data)
rows <- seq_len(nrow(data_tbl))
data_color_styles_tbl <- dplyr::tibble(locname = character(0),
grpname = character(0), colname = character(0), locnum = numeric(0),
rownum = integer(0), colnum = integer(0), styles = list())
for (i in seq_along(resolved_source_columns)) {
data_vals <- data_tbl[[resolved_source_columns[i]]][rows]
if (inherits(colors, "character")) {
if (is.numeric(data_vals)) {
color_fn <- scales::col_numeric(palette = colors,
domain = data_vals, alpha = TRUE)
}
else if (is.character(data_vals) || is.factor(data_vals)) {
if (length(colors) > 1) {
nlvl <- if (is.factor(data_vals)) {
nlevels(data_vals)
}
else {
nlevels(factor(data_vals))
}
if (length(colors) > nlvl) {
colors <- colors[seq_len(nlvl)]
}
}
color_fn <- scales::col_factor(palette = colors,
domain = data_vals, alpha = TRUE)
}
else {
cli::cli_abort("Don't know how to map colors to a column of class {class(data_vals)[1]}.")
}
}
else if (inherits(colors, "function")) {
color_fn <- colors
}
else {
cli::cli_abort("The `colors` arg must be either a character vector of colors or a function.")
}
color_fn <- rlang::eval_tidy(color_fn, data_tbl)
color_vals <- color_fn(data_vals)
color_vals <- html_color(colors = color_vals, alpha = alpha)
color_styles <- switch(apply_to, fill = lapply(color_vals,
FUN = function(x) cell_fill(color = x)), text = lapply(color_vals,
FUN = function(x) cell_text(color = x)))
data_color_styles_tbl <- dplyr::bind_rows(data_color_styles_tbl,
generate_data_color_styles_tbl(column = resolved_target_columns[i], rows = rows,
color_styles = color_styles))
if (apply_to == "fill" && autocolor_text) {
color_vals <- ideal_fgnd_color(bgnd_color = color_vals)
color_styles <- lapply(color_vals, FUN = function(x) cell_text(color = x))
data_color_styles_tbl <- dplyr::bind_rows(data_color_styles_tbl,
generate_data_color_styles_tbl(column = resolved_target_columns[i],
rows = rows, color_styles = color_styles))
}
}
dt_styles_set(data = data, styles = dplyr::bind_rows(dt_styles_get(data = data),
data_color_styles_tbl))
}
library(gt)
tmpfun <- get("data_color", envir = asNamespace("gt"))
environment(my_data_color) <- environment(tmpfun)
library(dplyr)
library(glue)
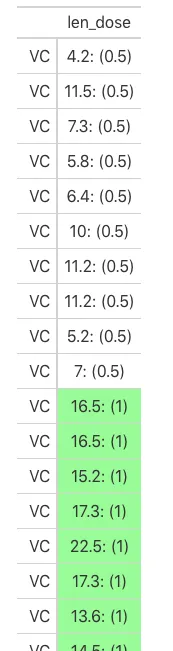
ToothGrowth %>%
mutate(len_dose = glue('{len}: ({dose})')) %>%
gt(rowname_col = 'supp') %>%
cols_hide(c(len, dose)) %>%
my_data_color(SOURCE_columns = "dose", TARGET_columns = "len_dose",
colors = scales::col_numeric(palette = c("red", "green"), domain = c(min(ToothGrowth$dose), max(ToothGrowth$dose))))
使用 reprex v2.0.2 工具于 2022-11-03 创建
data_color()函数。 - Lukas Wallrich