我刚开始使用 ggplot2 中的 geom_map 函数。在阅读了这里能找到的 29 篇关于 geom_map 的帖子之后,我仍然遇到了同样的问题。
我的数据框非常大,包含超过 2000 行。基本上,它是从世界卫生组织编制的有关特定基因(TP53)的数据。
请从这里下载。
标题如下:
> head(ARCTP53_SOExample)
Mutation_ID MUT_ID hg18_Chr17_coordinates hg19_Chr17_coordinates ExonIntron Genomic_nt Codon_number
1 16 1789 7519192 7578467 5-exon 12451 155
2 13 1741 7519200 7578475 5-exon 12443 152
3 17 2143 7519131 7578406 5-exon 12512 175
4 14 2143 7519131 7578406 5-exon 12512 175
5 15 2168 7519128 7578403 5-exon 12515 176
6 12 3737 7517845 7577120 8-exon 13798 273
Description c_description g_description g_description_hg18 WT_nucleotide Mutant_nucleotide
1 A>G c.463A>G g.7578467T>C NC_000017.9:g.7519192T>C A G
2 C>T c.455C>T g.7578475G>A NC_000017.9:g.7519200G>A C T
3 G>A c.524G>A g.7578406C>T NC_000017.9:g.7519131C>T G A
4 G>A c.524G>A g.7578406C>T NC_000017.9:g.7519131C>T G A
5 G>T c.527G>T g.7578403C>A NC_000017.9:g.7519128C>A G T
6 G>A c.818G>A g.7577120C>T NC_000017.9:g.7517845C>T G A
Splice_site CpG_site Type Mut_rate WT_codon Mutant_codon WT_AA Mutant_AA ProtDescription
1 no no A:T>G:C 0.170 ACC GCC Thr Ala p.T155A
2 no yes G:C>A:T at CpG 1.243 CCG CTG Pro Leu p.P152L
3 no yes G:C>A:T at CpG 1.280 CGC CAC Arg His p.R175H
4 no yes G:C>A:T at CpG 1.280 CGC CAC Arg His p.R175H
5 no no G:C>T:A 0.054 TGC TTC Cys Phe p.C176F
6 no yes G:C>A:T at CpG 1.335 CGT CAT Arg His p.R273H
Mut_rateAA Effect Structural_motif Putative_stop Sample_Name Sample_ID Sample_source Tumor_origin Grade
1 0.170 missense NDBL/beta-sheets 0 CAS91-19 17 surgery primary
2 1.243 missense NDBL/beta-sheets 0 CAS91-4 14 surgery primary
3 1.280 missense L2/L3 0 CAS91-13 12 surgery primary
4 1.280 missense L2/L3 0 CAS91-5 15 surgery primary
5 0.054 missense L2/L3 0 CAS91-1 16 surgery primary
6 1.335 missense L1/S/H2 0 CAS91-3 13 surgery primary
Stage TNM p53_IHC KRAS_status Other_mutations Other_associations
1 <NA> <NA> <NA>
2 <NA> <NA> <NA>
3 <NA> <NA> <NA>
4 <NA> <NA> <NA>
5 <NA> <NA> <NA>
6 <NA> <NA> <NA>
Add_Info Individual_ID Sex Age Ethnicity
1 Mutation only present in adjacent dysplastic area (Barrett's esophagus) 17 <NA> NA
2 Mutation only present in adjacent dysplastic area (Barrett's esophagus) 14 <NA> NA
3 Mutation only present in adjacent dysplastic area (Barrett's esophagus) 12 <NA> NA
4 Mutation only present in adjacent dysplastic area (Barrett's esophagus) 15 <NA> NA
5 16 <NA> NA
6 Mutation absent from adjacent dysplasia area (Barrett's esophagus) 13 <NA> NA
Geo_area Country Development Population Region TP53polymorphism Germline_mutation
1 USA More developed regions Northern America Americas NA
2 USA More developed regions Northern America Americas NA
3 USA More developed regions Northern America Americas NA
4 USA More developed regions Northern America Americas NA
5 USA More developed regions Northern America Americas NA
6 USA More developed regions Northern America Americas NA
Family_history Tobacco Alcohol Exposure Infectious_agent Ref_ID Cross_Ref_ID PubMed Exclude_analysis
1 <NA> <NA> <NA> <NA> 4 NA 1868473 False
2 <NA> <NA> <NA> <NA> 4 NA 1868473 False
3 <NA> <NA> <NA> <NA> 4 NA 1868473 False
4 <NA> <NA> <NA> <NA> 4 NA 1868473 False
5 <NA> <NA> <NA> <NA> 4 NA 1868473 False
6 <NA> <NA> <NA> <NA> 4 NA 1868473 False
WGS_WXS
1 No
2 No
3 No
4 No
5 No
6 No
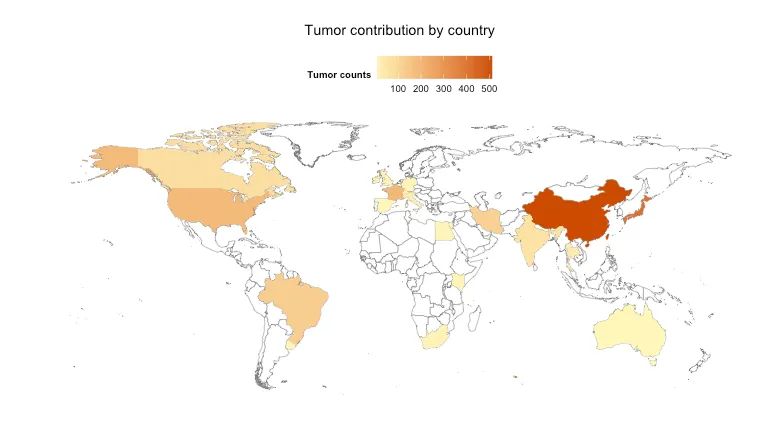
无论如何,我想创建一个简单的世界地图,着重标注进行过这种突变研究的国家,并在这些国家中有更多或更少的“突变特征”。如果您能看到这个链接,您可能会更好地理解我的意图:
summary(ARCTP53_SOExample$Country)
Australia Brazil Canada China
1 127 76 519
China, Hong-Kong Chinese Taipei (Taiwan) Czech Republic Egypt
52 36 9 9
France Germany India Iran
195 10 63 112
Ireland Italy Japan Kenya
25 30 414 11
South Africa Spain Switzerland Thailand
13 2 24 35
The Netherlands UK Uruguay USA
6 17 6 189
NA's
30
我的数据框中有一些国家出现了多次。
所以为了获得我想要的地图,我做了以下操作:
library(ggplot2)
library(maps)
world_map<-map_data("world")
ggplot(ARCTP53_SOExample)+geom_map(map = world_map, aes(map_id = Country,fill = Country),
+ colour = "black") +
+ expand_limits(x = world_map$long, y = world_map$lat)
这是我得到的结果:
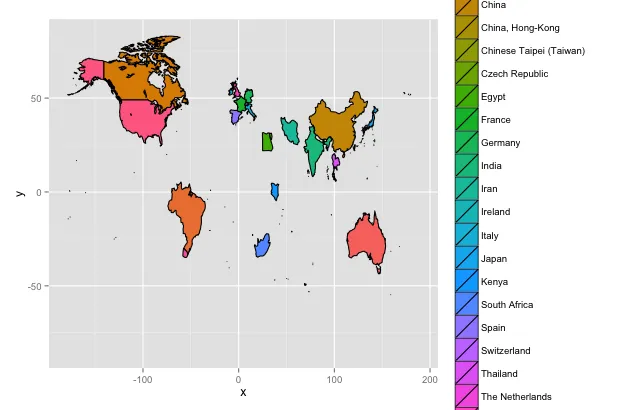 。请问有人能提供我做错了什么吗?
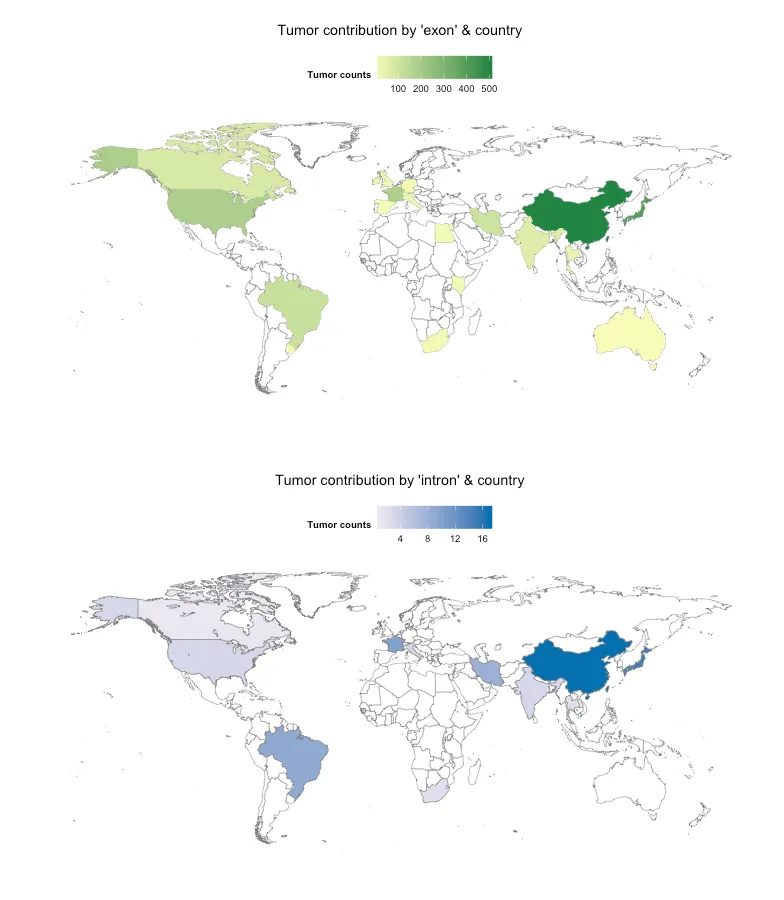
。请问有人能提供我做错了什么吗?此外,未来我想在不同国家之间添加ExonIntron列的geom_bar()。但首先我想尝试生成正确的地图。谢谢!
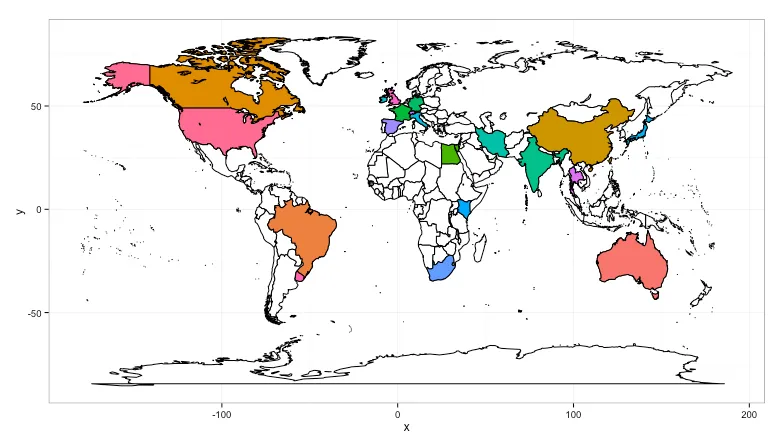
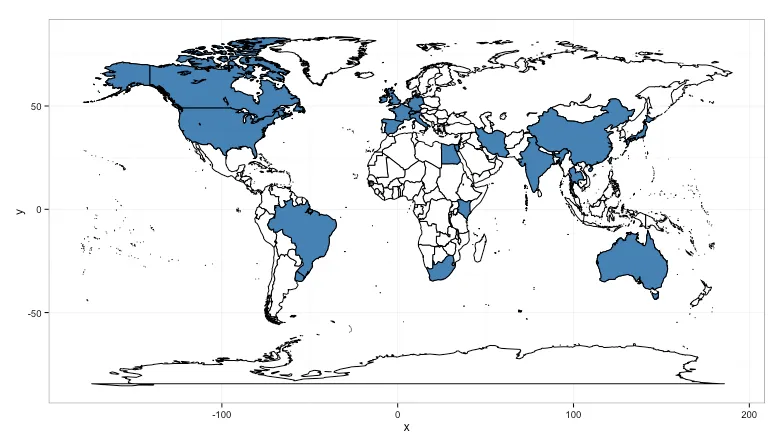
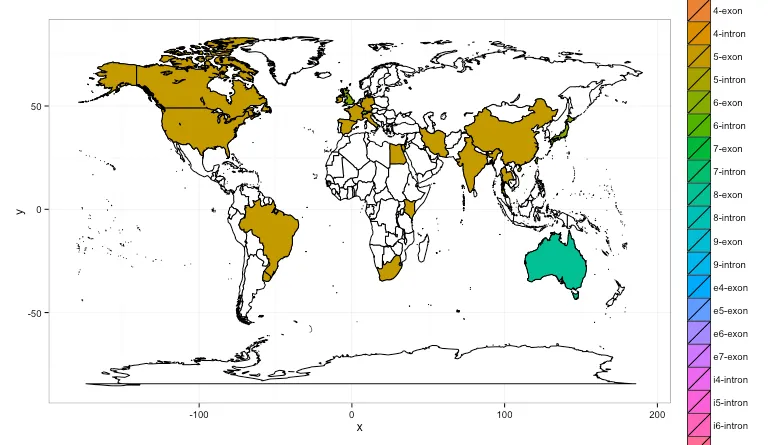
gridExtra包中。我将代码放在了github上,以便更容易地获取:https://gist.github.com/hrbrmstr/0a330b212b46517ffee9,并包含了一个片段,展示如何查看错误的区域名称。您可以使用基本R包中的`aggregate`而不是`plyr`中的`count`,但我认为`count`语法更清晰。 - hrbrmstr.(region)语法是plyr包中用于指定列名的语法糖。它等同于c("region")(而且c()不是必需的,但这是我的习惯)。您可以随时通过 bob at rudis dot net 联系我获取更多信息(在 SO 上进行后续操作有点困难,反正我很乐意帮忙)。 - hrbrmstr