我编写了一个特殊的“impute”函数,根据特定列名,用mean()或mode()替换具有缺失(NA)值的列值。
输入数据框有400,000+行,速度非常慢,如何使用lapply()或apply()加速填充部分。
这是函数,我想要优化的部分用START OPTIMIZE和END OPTIMIZE标记:
specialImpute <- function(inputDF)
{
discoveredDf <- data.frame(STUDYID_SUBJID=character(), stringsAsFactors=FALSE)
dfList <- list()
counter = 1;
Whilecounter = nrow(inputDF)
#for testing just do 10 iterations,i = 10;
while (Whilecounter >0)
{
studyid_subjid=inputDF[Whilecounter,"STUDYID_SUBJID"]
vect = which(discoveredDf$STUDYID_SUBJID == studyid_subjid)
#was discovered and subset before
if (!is.null(vect))
{
#not subset before
if (length(vect)<1)
{
#subset the dataframe base on regex inputDF$STUDYID_SUBJID
df <- subset(inputDF, regexpr(studyid_subjid, inputDF$STUDYID_SUBJID) > 0)
#START OPTIMIZE
for (i in nrow(df))
{
#impute , add column mean & add to list
#apply(df[,c("y1","y2","y3","etc..")],2,function(x){x[is.na(x)] =mean(x, na.rm=TRUE)})
if (is.na(df[i,"y1"])) {df[i,"y1"] = mean(df[,"y1"], na.rm = TRUE)}
if (is.na(df[i,"y2"])) {df[i,"y2"] =mean(df[,"y2"], na.rm = TRUE)}
if (is.na(df[i,"y3"])) {df[i,"y3"] =mean(df[,"y3"], na.rm = TRUE)}
#impute using mean for CONTINUOUS variables
if (is.na(df[i,"COVAR_CONTINUOUS_2"])) {df[i,"COVAR_CONTINUOUS_2"] =mean(df[,"COVAR_CONTINUOUS_2"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_3"])) {df[i,"COVAR_CONTINUOUS_3"] =mean(df[,"COVAR_CONTINUOUS_3"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_4"])) {df[i,"COVAR_CONTINUOUS_4"] =mean(df[,"COVAR_CONTINUOUS_4"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_5"])) {df[i,"COVAR_CONTINUOUS_5"] =mean(df[,"COVAR_CONTINUOUS_5"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_6"])) {df[i,"COVAR_CONTINUOUS_6"] =mean(df[,"COVAR_CONTINUOUS_6"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_7"])) {df[i,"COVAR_CONTINUOUS_7"] =mean(df[,"COVAR_CONTINUOUS_7"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_10"])) {df[i,"COVAR_CONTINUOUS_10"] =mean(df[,"COVAR_CONTINUOUS_10"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_14"])) {df[i,"COVAR_CONTINUOUS_14"] =mean(df[,"COVAR_CONTINUOUS_14"], na.rm = TRUE)}
if (is.na(df[i,"COVAR_CONTINUOUS_30"])) {df[i,"COVAR_CONTINUOUS_30"] =mean(df[,"COVAR_CONTINUOUS_30"], na.rm = TRUE)}
#impute using mode ordinal & nominal values
if (is.na(df[i,"COVAR_ORDINAL_1"])) {df[i,"COVAR_ORDINAL_1"] =Mode(df[,"COVAR_ORDINAL_1"])}
if (is.na(df[i,"COVAR_ORDINAL_2"])) {df[i,"COVAR_ORDINAL_2"] =Mode(df[,"COVAR_ORDINAL_2"])}
if (is.na(df[i,"COVAR_ORDINAL_3"])) {df[i,"COVAR_ORDINAL_3"] =Mode(df[,"COVAR_ORDINAL_3"])}
if (is.na(df[i,"COVAR_ORDINAL_4"])) {df[i,"COVAR_ORDINAL_4"] =Mode(df[,"COVAR_ORDINAL_4"])}
#nominal
if (is.na(df[i,"COVAR_NOMINAL_1"])) {df[i,"COVAR_NOMINAL_1"] =Mode(df[,"COVAR_NOMINAL_1"])}
if (is.na(df[i,"COVAR_NOMINAL_2"])) {df[i,"COVAR_NOMINAL_2"] =Mode(df[,"COVAR_NOMINAL_2"])}
if (is.na(df[i,"COVAR_NOMINAL_3"])) {df[i,"COVAR_NOMINAL_3"] =Mode(df[,"COVAR_NOMINAL_3"])}
if (is.na(df[i,"COVAR_NOMINAL_4"])) {df[i,"COVAR_NOMINAL_4"] =Mode(df[,"COVAR_NOMINAL_4"])}
if (is.na(df[i,"COVAR_NOMINAL_5"])) {df[i,"COVAR_NOMINAL_5"] =Mode(df[,"COVAR_NOMINAL_5"])}
if (is.na(df[i,"COVAR_NOMINAL_6"])) {df[i,"COVAR_NOMINAL_6"] =Mode(df[,"COVAR_NOMINAL_6"])}
if (is.na(df[i,"COVAR_NOMINAL_7"])) {df[i,"COVAR_NOMINAL_7"] =Mode(df[,"COVAR_NOMINAL_7"])}
if (is.na(df[i,"COVAR_NOMINAL_8"])) {df[i,"COVAR_NOMINAL_8"] =Mode(df[,"COVAR_NOMINAL_8"])}
}#for
#END OPTIMIZE
dfList[[counter]] <- df
#add to discoveredDf since already substed
discoveredDf[nrow(discoveredDf)+1,]<- c(studyid_subjid)
counter = counter +1;
#for debugging to check progress
if (counter %% 100 == 0)
{
print(counter)
}
}
}
Whilecounter = Whilecounter -1;
}#end while
return (dfList)
}
谢谢
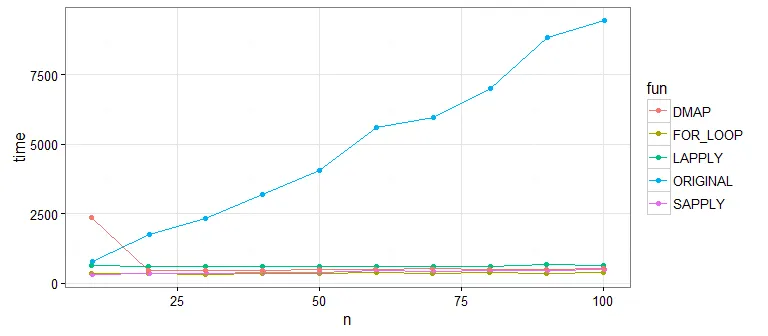
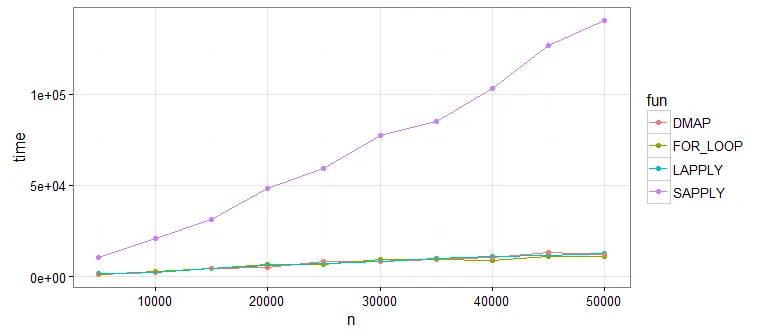
if (is.na(df[i, "y1"] ...移到循环外面,作为df[is.na(df$y1), "y1"] = mean(df$y1, na.rm=TRUE)。这样可以将迭代向量化,每列只有一个 R 调用,而不是 nrow(df) 次调用。对循环中的所有行重复此操作。 - Martin Morgan