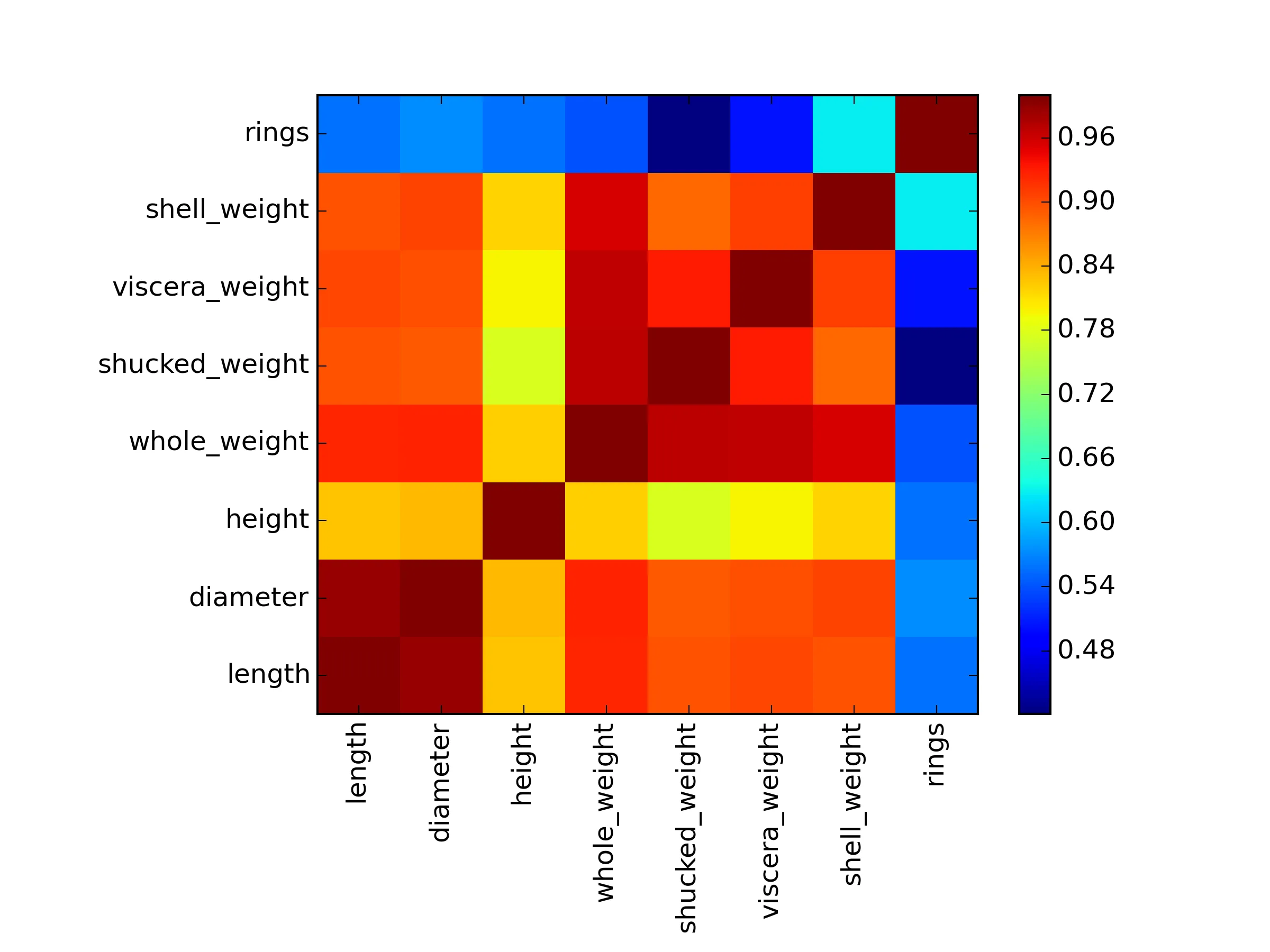
DF_correlation = [[ 1. 0.98681158 0.82755361 0.92526117 0.89791366 0.9030177
0.89770557 0.55671958]
[ 0.98681158 1. 0.83368369 0.9254521 0.89316248 0.89972443
0.90532978 0.57465985]
[ 0.82755361 0.83368369 1. 0.81922077 0.77497229 0.7983193
0.81733801 0.55746732]
[ 0.92526117 0.9254521 0.81922077 1. 0.96940546 0.96637508
0.95535544 0.54038968]
[ 0.89791366 0.89316248 0.77497229 0.96940546 1. 0.93196132
0.88261706 0.42088366]
[ 0.9030177 0.89972443 0.7983193 0.96637508 0.93196132 1.
0.90765632 0.50381925]
[ 0.89770557 0.90532978 0.81733801 0.95535544 0.88261706 0.90765632
1. 0.62757404]
[ 0.55671958 0.57465985 0.55746732 0.54038968 0.42088366 0.50381925
0.62757404 1. ]]
我正在按照https://www.geekbooks.me/book/view/machine-learning-in-python的指导,制作回归热力图。
import pandas as pd
from pandas import DataFrame
import matplotlib.pyplot as plt
headers = ["sex", "length","diameter", "height", "whole_weight", "shucked_weight","viscera_weight","shell_weight","rings"]
以下是Michael Bowles的代码:
plt.pcolor(DF_correlation)
plt.show()
这段代码可以正常工作,但是没有标签。为了加入标签,我尝试了matplotlib: colorbars and its text labels中的方法。虽然我稍微修改了一下格式,但还是没有成功。
fig, ax = plt.subplots()
heatmap = ax.pcolor(DF_correlation)
cbar = plt.colorbar(heatmap)
ax.set_xticklabels = ax.set_yticklabels = headers[1:]
plt.show()
如何给这个图添加标签?由于这是一个相关性图,因此x轴和y轴的标签将是相同的......基本上是 headers[1:]