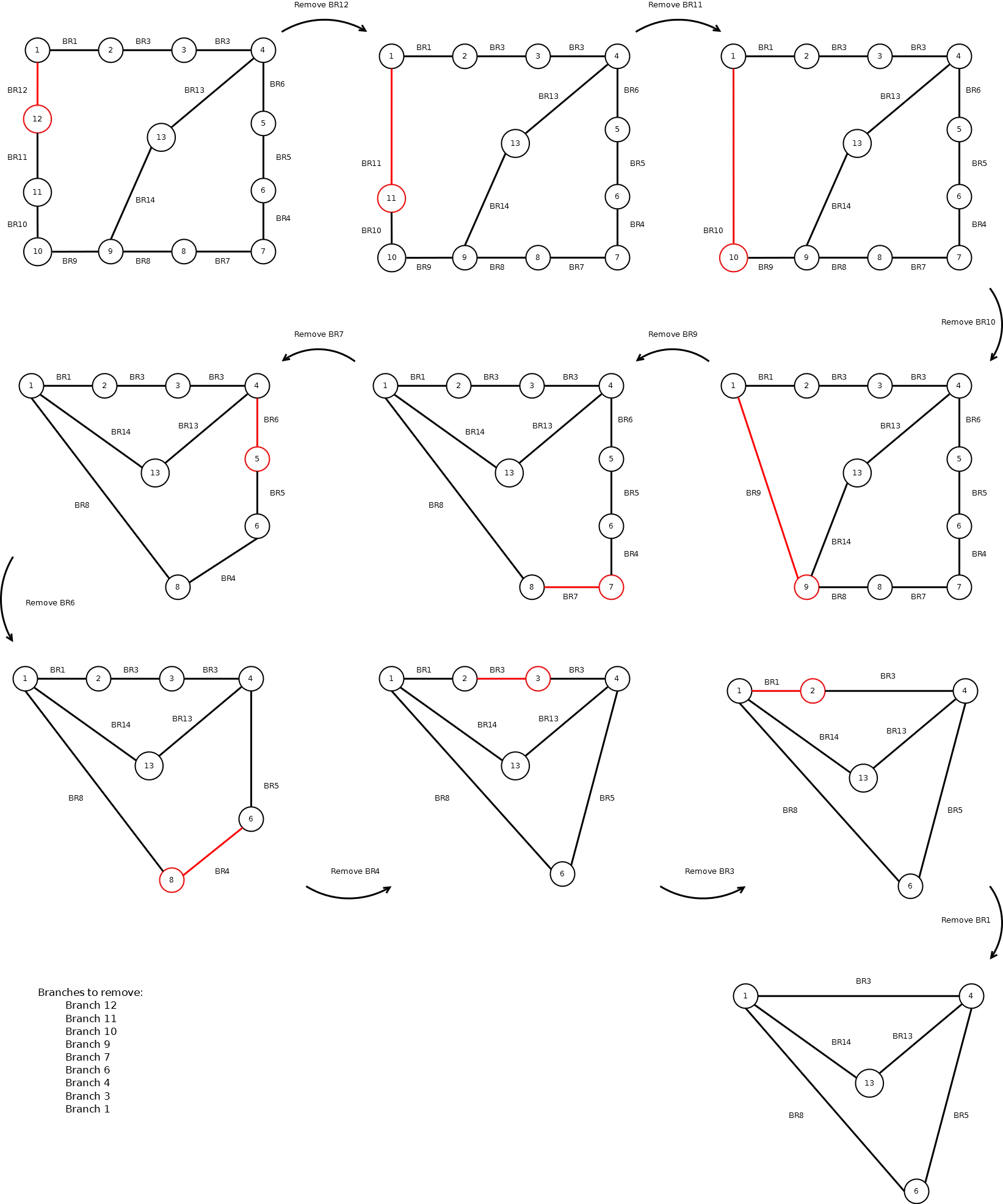
我一直在编写代码以减少一个图形。问题是有些分支我想要移除。一旦我移除了一个分支,我可以合并节点或不合并,这取决于分支连接的节点之间路径的数量。
也许以下示例说明了我的意思:
也许以下示例说明了我的意思:
我拥有的代码如下:
from networkx import DiGraph, all_simple_paths, draw
from matplotlib import pyplot as plt
# data preparation
branches = [(2,1), (3,2), (4,3), (4,13), (7,6), (6,5), (5,4),
(8,7), (9,8), (9,10), (10,11), (11,12), (12,1), (13,9)]
branches_to_remove_idx = [11, 10, 9, 8, 6, 5, 3, 2, 0]
ft_dict = dict()
graph = DiGraph()
for i, br in enumerate(branches):
graph.add_edge(br[0], br[1])
ft_dict[i] = (br[0], br[1])
# Processing -----------------------------------------------------
for idx in branches_to_remove_idx:
# get the nodes that define the edge to remove
f, t = ft_dict[idx]
# get the number of paths from 'f' to 't'
n_paths = len(list(all_simple_paths(graph, f, t)))
if n_paths == 1:
# remove branch and merge the nodes 'f' and 't'
#
# This is what I have no clue how to do
#
pass
else:
# remove the branch and that's it
graph.remove_edge(f, t)
print('Simple removal of', f, t)
# -----------------------------------------------------------------
draw(graph, with_labels=True)
plt.show()
我认为应该有一种更简单直接的方法,通过分支索引获取第一个数字的最后一个数字,但我毫无头绪。