你需要一个最短路径的矩阵,然后使用这些路径上所有边的并集创建子图。
将
关键顶点定义为所需子图出现的顶点。你说你有三个这样的关键顶点。
考虑到它们之间的任何
i和
j的最短路径是
unlist(shortest_paths(g, i, j, mode="all", weights=NULL)$vpath)。你需要列出所有i-j组合(在你的情况下是1-2、1-3、2-3),然后列出它们之间出现的所有顶点。有时候,同一个顶点会出现在多个ij对的最短路径上(参见
介数中心性)。你所需的子图应仅包括这些顶点,你可以将它们提供给
induced_subgraph()。
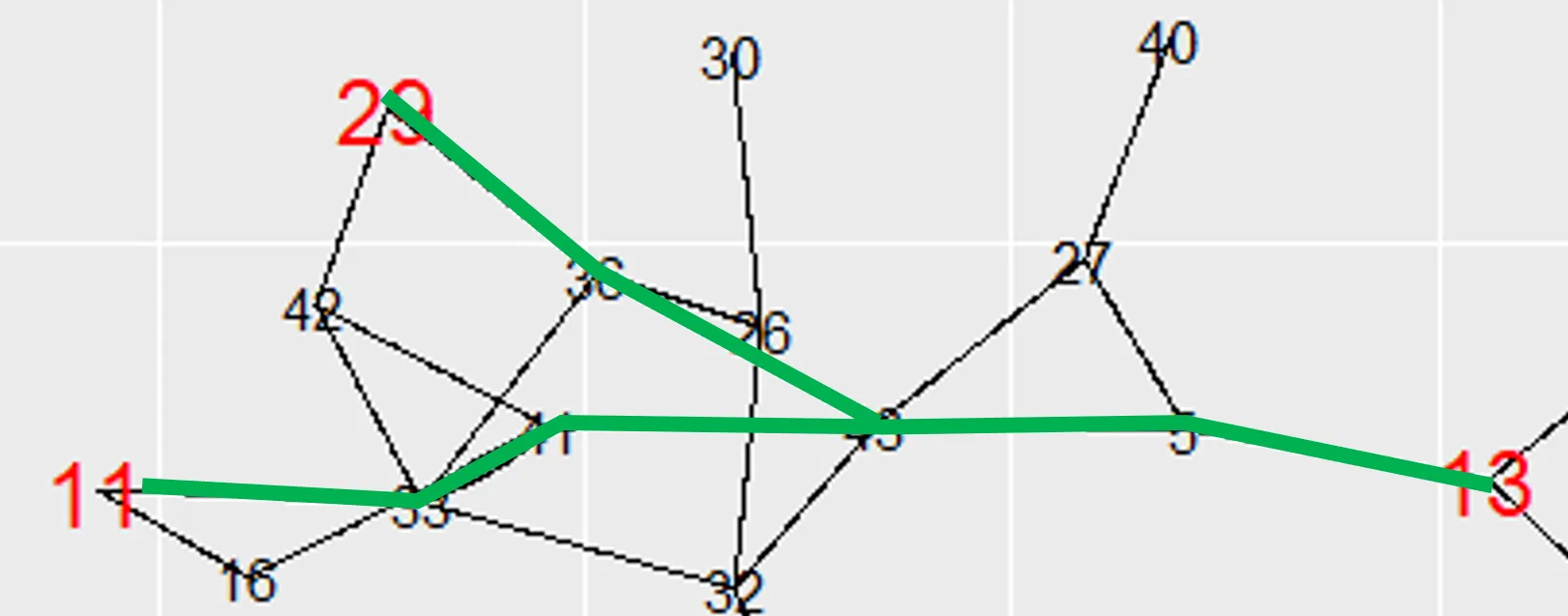
然后出现另一个有趣的问题。并非所有连接您选择的顶点的边都是最短路径的一部分。我不确定您在子图中需要什么,但我假设您只想要最短路径的顶点和边。
induced_subgraph() 的手册说明可以提供
eids来过滤边缘子图,但我无法使其起作用。如果有人解决了这个问题,欢迎发表评论。为了创建仅包含最短路径上实际的边和顶点的子图,必须删除一些多余的边。
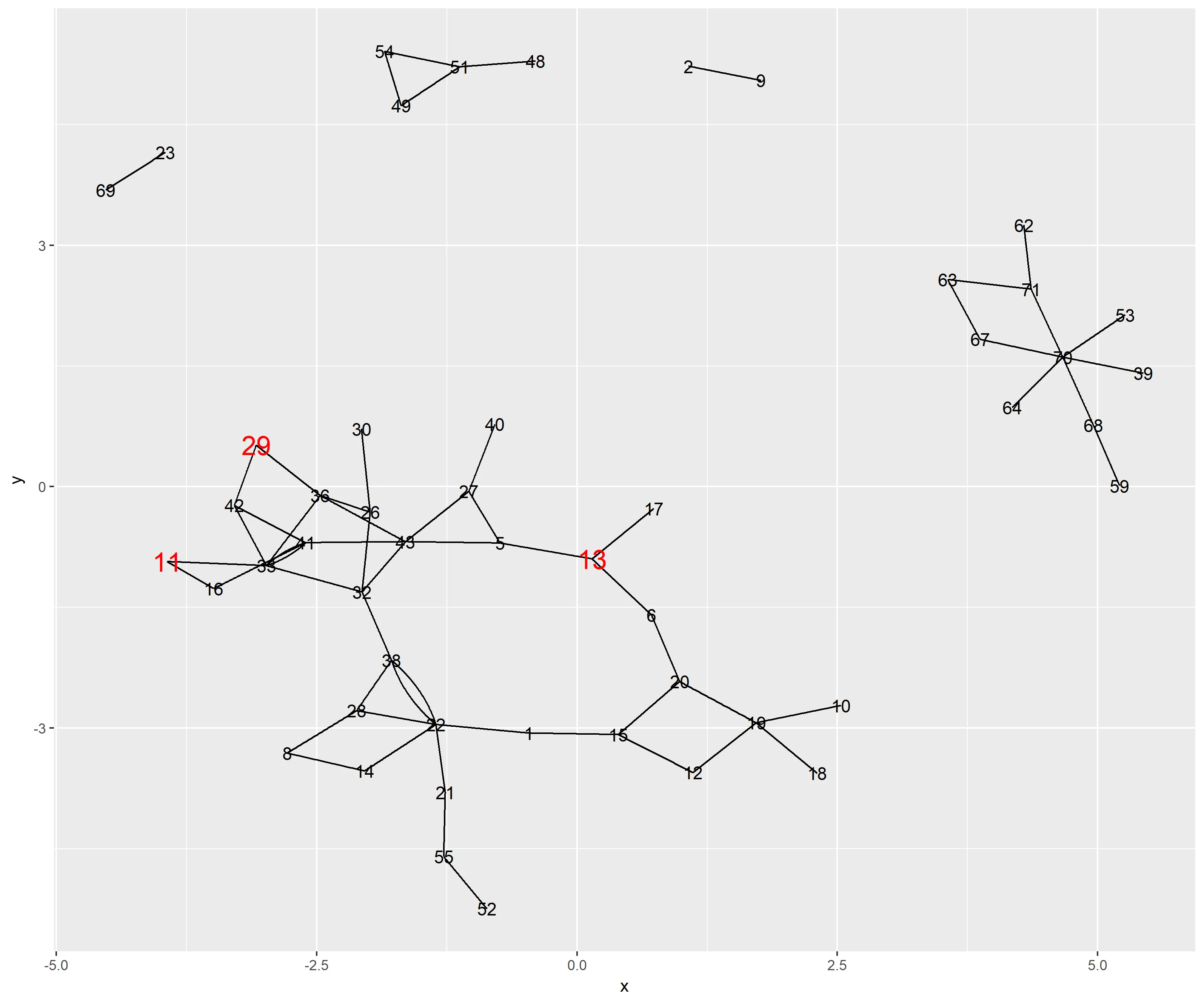
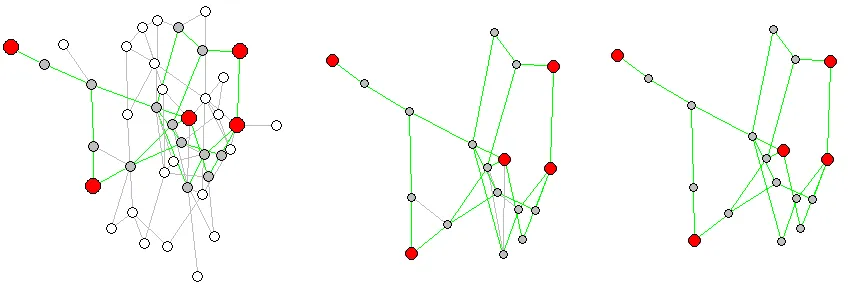
下面是一个示例,其中随机选择了一些关键顶点,可视化了子图的多余边问题,并生成了一个适当的仅限于更短路径的子图:
library(igraph)
N <- 40
E <- 70
K <- 5
g <- erdos.renyi.game(N, E, type="gnm", directed = FALSE, loops = FALSE)
V(g)$label <- NA
V(g)$color <- "white"
V(g)$size <- 8
E(g)$color <- "gray"
key_vertices <- sample(1:N, 5)
g <- g %>% set_vertex_attr("color", index=key_vertices, value="red")
g <- g %>% set_vertex_attr("size", index=key_vertices, value=12)
get_path <- function(x){
i <- x[1]; j <- x[2]
if(distances(g,i,j) == Inf){
path <- c()
} else {
path <- unlist(shortest_paths(g, i, j, mode="all", weights=NULL)$vpath)
}
}
key_el <- expand.grid(key_vertices, key_vertices)
key_el <- key_el[key_el$Var1 != key_el$Var2,]
paths <- apply(key_el, 1, get_path)
path_vertices <- setdiff(unique(unlist(paths)), key_vertices)
g <- g %>% set_vertex_attr("color", index=path_vertices, value="gray")
mark_edges <- function(path, edges=c()){
for(n in 1:(length(path)-1)){
i <- path[n]
j <- path[1+n]
edge <- get.edge.ids(g, c(i,j), directed = TRUE, error=FALSE, multi=FALSE)
edges <- c(edges, edge)
}
(edges)
}
key_edges <- lapply(paths, function(x) if(length(x) > 1){mark_edges(x)})
key_edges <- unique(unlist(key_edges))
g <- g %>% set_edge_attr("color", index=key_edges, value="green")
plot(g)
sg_vertices <- sort(union(key_vertices, path_vertices))
unclean_sg <- induced_subgraph(g, sg_vertices)
plot(unclean_sg)
sg <- delete.edges(unclean_sg, which(E(unclean_sg)$color=="gray"))
l <-layout.auto(g)
layout(matrix(c(1,1,2,3), 2, 2, byrow = TRUE))
plot(g, layout=l)
plot(unclean_sg, layout=l[sg_vertices,])
plot(sg, layout=l[sg_vertices,])